D31.3 Enzymes
Millions of chemical reactions occur within each of our cells every minute, and many of these reactions would proceed at an extremely slow rate without catalysts. For example, the dissociation of carbonic acid that takes place in the lungs:
only proceeds at a rate of ~10-7 M/s at room temperature. However, CO2 needs to be produced at a much higher rate than this for the required quantity to be exhaled every minute. While it is theoretically possible to accelerate the reaction by raising the temperature, unfortunately, most life is only compatible with a limited temperature range.
Biological catalysts, known as enzymes, are crucial for life because they can accelerate reactions by factors of 106-1020. For example, the enzyme carbonic anhydrase accelerates the above reaction at room temperature by more than a million times.
Like all catalysts, enzymes act by combining with reactants and thereby forming lower energy transition states. In an enzyme-catalyzed reaction, the reactant(s) with which the enzyme combines is(are) called the substrate(s). Unlike chemical catalysts, an enzyme’s interactions with substrate molecules are often partly or entirely noncovalent; that is, the interactions involve hydrogen bonding, ionic attractions, and/or dipole-dipole attractions.
In addition, enzyme-catalyzed reactions are highly specific to a particular substrate or a particular category of substrate molecules. Enzymes are said to “recognize” the substrate for which the enzyme serves as a catalyst. Enzyme interactions with substrates are sometimes so specific that an enzyme does not recognize a molecule that differs from its preferred substrate by as little as a single methyl (-CH3) group, and many enzymes will recognize one enantiomer but not its mirror image.
How do enzymes accelerate chemical reactions and achieve their specificity? The answer to both questions lies in how enzymes interact with their substrates. Typically, an enzyme is a protein molecule. Usually only part of the large enzyme molecule interacts with a substrate, and this part is called the active site.
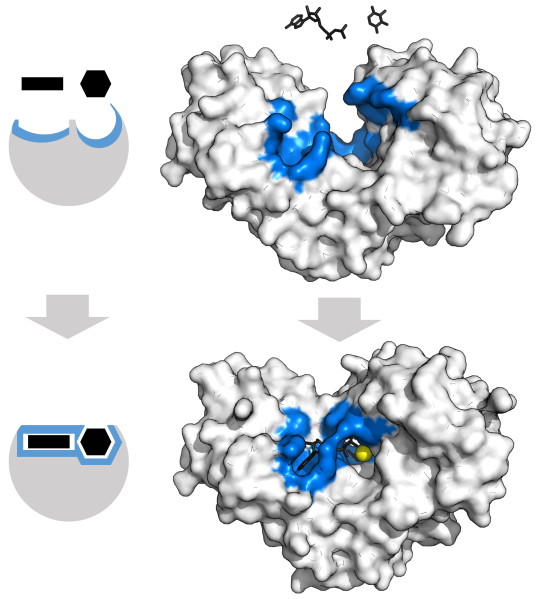
In 1890, the chemist Emil Fischer proposed that the substrate fits into the enzyme’s active site as a key fits into a lock. The key (substrate) has a specific molecular shape (arrangement of functional groups and other atoms) that allows it, and no other key, to fit into the lock (the enzyme’s active site).
In 1958, Daniel E. Koshland Jr. modified this lock-and-key model by proposing that binding of the substrate to the enzyme alters the configuration of both, providing a better fit. This induced-fit model explains both the large increase in reaction rates and the specificity of enzyme-catalyzed reactions:
- As the substrate interacts with the enzyme, the substrate is distorted (atoms are shifted, bonds are stretched, and reactive groups are brought close together) to a structure closer to the transition state of the reaction. This lowers the energy of the transition state, accelerating the reaction.
- Only molecules with the correct functional groups in the correct configurations are able to be induced to fit the active site of the enzyme.
Please use this form to report any inconsistencies, errors, or other things you would like to change about this page. We appreciate your comments. 🙂 (Note that we cannot answer questions via the google form. If you have a question, please post it on Piazza.)

