Lab 4 Background
Determination of DNA Concentration and Purity Using a Spectrophotometer
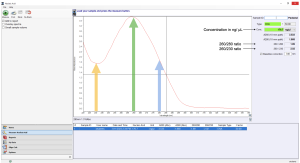
A spectrophotometer measures the intensity of the light beam at a different wavelength. It can be used to measure the concentration and purity of DNA because DNA exhibits a characteristic light absorbance pattern. For a pure DNA sample, it absorbs a maximum amount of UV light at 260nm, and it absorbs about half as much UV light at 280nm. In this lab, we will use a spectrophotometer called a NanoDrop (manufacture name) to measure the absorbance at 260 nm, or A260, of your purified DNA samples. The spectrophotometer will simultaneously measure the absorbance at 280nm, and pure DNA should give a 260/280 ratio of 1.8.
The NanoDrop will also simultaneously measure the absorbance at 230nm, which shows small molecule contaminations in the DNA sample. The expected A260/A230 value for pure DNA is in the range of 2 and 2.2; a lower ratio indicates the presence of containments in the sample.
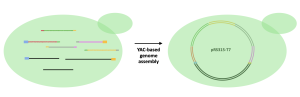
Yeast Artificial Chromosome (YAC)-Based Genome Assembly
YAC is a genetically engineered chromosome originated from yeast and can be used to assemble and amplify DNA in yeast cells. To accomplish a YAC assembly, one or more DNA fragments and the YAC need to be taken up by yeast cells using various methods. In our case, a method called transformation will be used to “insert” DNA fragments into yeast cells. Once inside, yeast cells identify the features on the fragments and assemble them into one circular DNA. Because YAC mimics endogenous yeast chromosomes, yeast cells will assemble and amplify the inserted DNA (in other words, yeast cannot distinguish its own chromosomes and YAC, and will replicate YAC as if the YAC is the yeast’s own chromosome). The amplified DNA will then be extracted from yeast cells for later use.
In the Biochem375 laboratory, we will put all PCR amplified fragments, including both the T7 genome fragments, and the YAC fragments into yeast cells. Yeast cells will then assemble all the fragments in the correct order into one vector (circular DNA) and then amplify the T7 genome, as shown in the figure below.
Reference
Ando, H., Lemire, S., Pires, D. P., & Lu, T. K. (2015). Engineering Modular Viral Scaffolds for Targeted Bacterial Population Editing. Cell Syst, 1(3), 187-196. doi:10.1016/j.cels.2015.08.013
Huss, P., Meger, A., Leander, M., Nishikawa, K., & Raman, S. (2021). Mapping the functional landscape of the receptor binding domain of T7 bacteriophage by deep mutational scanning. Elife, 10. doi:10.7554/eLife.63775
