Introduction to Engineering Bacteriophage Laboratory
What are bacteriophages and phage therapy?
Bacteriophages, or phages, are viruses that can infect and kill bacteria. Depending on the species, phages can take on various shapes and can have either DNA or RNA as genetic material. Phages, or phage particles, are composed of genetic material and a protective protein shell called a capsid. A typical bacteriophage particle is shown below (Figure 1).
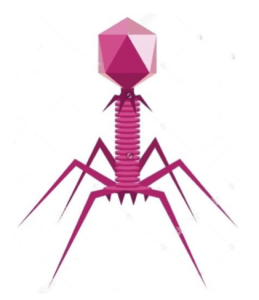
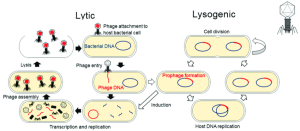
A phage particle can interact with the host cell (bacterial cell) by specific interaction between the phage proteins and host molecules. This interaction allows the phage to inject its genetic material into the target bacterial cell. Once inside, the phage’s genetic material has two fates, depending on whether a phage undergoes a lytic cycle or a lysogenic cycle (Figure 2).
In a lytic cycle, the genetic material is replicated and is also used to produce proteins to make new phage particles. The newly formed phage particles leave the host cells by breaking out the host’s cell membrane (lysing), killing the host in the process. In a lysogenic cycle, the genetic material of the phage is integrated into the bacterial genome and remains inactive for a period of time. Depending on environmental conditions and the phage type, the phage genome can remain integrated in the host genomes from minutes to years. In this stage, a phage is called a prophage as there is no viral protein production or phage particle formation. Under the right condition, the prophage becomes active and initiates the lytic cycle, leading to genetic material replication, protein production, phage particle formation and host cell lysis (Figure 2).
Because of its ability to infect and kill bacteria, phage therapy has been considered a natural treatment option for bacterial infections. Unlike mammalian-targeting viruses such as SARS-COVID 2, bacteriophage does not infect eukaryotic species and is thus considered a low-risk option to treat antibiotic resistance.
Antibiotics are a class of drugs that kill or inhibit the growth of bacteria. Since the discovery of penicillin by the physician and microbiologist Sir Alexander Fleming, antibiotics have saved many from deadly bacterial infections. However, overuse and misuse of antibiotics on humans and other animals exerted a strong evolutionary selection pressure on bacteria, and an increasing number of bacterial species are resistant to multiple, if not all known antibiotics. These “superbugs” pose a substantial threat to human health.
One potential regimen of antibiotic resistance that has gained increasing attention in recent years is phage therapy, which is a tailor-made approach that uses phage to kill disease-causing bacteria. While phage therapy has been used in Eastern Europe for decades, other countries have been slow in adopting phage therapy, in part due to the prominence and effectiveness of antibiotics. However, given the rise of antibiotic resistance, phage therapy becomes an attractive strategy, not least because of its specificity (does not target humans or other non-target bacteria) and effectiveness.
Bacteriophages are one of the most abundant living entities on earth, with many species that target various types of bacteria. A bacteriophage, depending on its type, can attack one, a few types of bacteria. On one hand, this specificity is highly desirable because it allows the specific killing of only the targeted bacteria and spare non-harmful or beneficial bacteria such as the gut microbiota. On the other hand, using phage therapy requires the identification of the harmful bacteria species, which can be unavailable in many cases. If the type of infectious bacteria is unknown, however, broad-spectrum phage curtail that contain many types of phages may need to be used which may lead to the killing of beneficial bacteria.
However, even in cases where the harmful bacteria species is not identified, phage therapy may still be preferred since bacteria is less likely to develop resistance to phage therapy than traditional antibiotic treatment. This is because, unlike antibiotics that have fixed chemical structures, the genetic material of phages can be, and indeed are, changing at a rate faster than bacteria. Thus, it is highly likely that the phage will adapt to and outwit bacteria defensive mechanisms, allowing it to continue to infect and kill bacteria.
How to engineer a T7 bacteriophage?
The genetic information of organisms can be altered through technology via a process called genetic engineering. Genetic engineering may involve changing one or more nucleotides of a gene/genes and adding or removing nucleotides from one or more regions of a gene/genes. Genetic engineering is usually done in two ways. One method is to isolate the genes or genome of the target organism, making genetic changes on the isolated genetic information, and puting the engineered genetic information back into the same or a different organism. In recent years, advanced technology, such as the CRISPR/Cas9 technology, also allows scientists to make changes directly on the genetic information within the organism without the isolation step. In this course, we will use the former technology to accomplish the T7 bacteriophage engineering.
What do we do in this laboratory?
In this laboratory, you will conduct hypothesis-driven, real-life research to create and to understand the virulence of T7 bacteriophage genetic variants on various bacterial strains. Specifically, you will manipulate the DNA of a region on the T7 bacteriophage genome that is known to interact and allow the entry of the phage into the host. By genetically engineering the genetic information of that region, you will investigate the effect of the mutation on the virulence of the T7 phage, and gain a better understanding of how the T7 bacteriophage infects bacteria.
In the process, you will learn state-of-the-art techniques that are routinely used in biochemistry and microbiology laboratories in industry and academia. In the project we will follow the outline below:
- Identify the region of T7 that is critical to the phage-bacteria interaction
- Choose a mutation of the T7 phage based on amino acid structure and the protein structure
- Construct the T7 phage harboring the mutant using YAC-based genome assembly
- Express the phage in bacteria to procure the mutant T7 phage
- Enumerate the mutant phage
- Test the virulence of the wild-type and the mutant phages on one type of bacteria host
- Test the virulence of the wild-type and mutant phages in multiple types of bacteria hosts
- Investigate the effect of phage treatment on a limited bacterial ecosystem
Reference
Ando, H., Lemire, S., Pires, D. P., & Lu, T. K. (2015). Engineering Modular Viral Scaffolds for Targeted Bacterial Population Editing. Cell Syst, 1(3), 187-196. doi:10.1016/j.cels.2015.08.013
Batinovic, S., Wassef, F., Knowler, S. A., Rice, D. T. F., Stanton, C. R., Rose, J., . . . Franks, A. E. (2019). Bacteriophages in Natural and Artificial Environments. Pathogens, 8(3). doi:10.3390/pathogens8030100
Dunn, J. J., & Studier, F. W. (1983). Complete nucleotide sequence of bacteriophage T7 DNA and the locations of T7 genetic elements. J Mol Biol, 166(4), 477-535. doi:10.1016/s0022-2836(83)80282-4
Duzgunes, N., Sessevmez, M., & Yildirim, M. (2021). Bacteriophage Therapy of Bacterial Infections: The Rediscovered Frontier. Pharmaceuticals (Basel), 14(1). doi:10.3390/ph14010034
Huss, P., Meger, A., Leander, M., Nishikawa, K., & Raman, S. (2021). Mapping the functional landscape of the receptor binding domain of T7 bacteriophage by deep mutational scanning. Elife, 10. doi:10.7554/eLife.63775
Hyman, P. (2019). Phages for Phage Therapy: Isolation, Characterization, and Host Range Breadth. Pharmaceuticals (Basel), 12(1). doi:10.3390/ph12010035
Squires, R. A. (2021). Bacteriophage therapy for challenging bacterial infections: achievements, limitations and prospects for future clinical use by veterinary dermatologists. Vet Dermatol, 32(6), 587-e158. doi:10.1111/vde.12958
