3 Structure of a Scientific Research Paper
A primary way that scientists communicate with one another is through scientific papers. We will model our Biocore lab reports on the format most commonly used by scientific journals. Your lab reports should follow the guidelines described below unless the lab manual or your TA specifically tells you otherwise. Some lab reports have a modified format or require only a subset of the standard sections listed below.
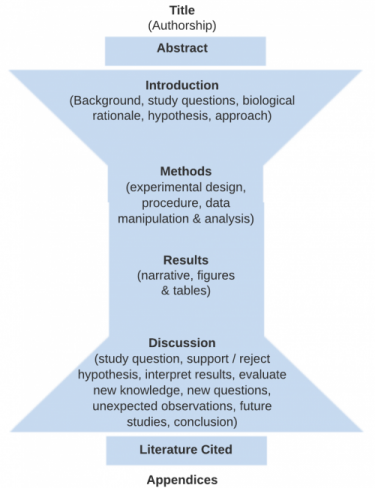
The figure below indicates the four main sections (Intro, Methods, Results and Discussion) that form the body of a scientific paper. Each section of the paper (except for “Title”) should begin with one of these terms as a heading These main sections are bookended on the front end with a Title and Abstract summarizing the whole document and on the back end by a Literature Cited and Appendices (optional) in support of the document.
Other classes and some scientific journals deviate from this format, and you should always consult the guidelines specified before preparing a paper for another class (or submitting a manuscript for publication).
- Title
- Authorship
- Abstract
- Introduction
- Methods and Materials
- Results (including figures and tables)
- Discussion
- Literature Cited
The Methods and Results are specific to your hypothesis and the experiment you performed.
Then the Discussion starts more narrowly focused on whether you support or reject your hypothesis, but then broadens to integrate your findings into the existing literature, and finishes with a conclusion that is based on the experimental evidence you present.
Title
The title is a clear, specific statement of the subject of your report. Think of the words in your title as key search terms. It introduces the reader to your paper and lets them know what to expect.
Titles should:
- Be concise and informative and need not be complete sentences.
- Avoid filler words like “Studies on” or “Investigations of” and opening words like A, An, or The.
- Be as specific as possible.
- Avoid abbreviations and jargon.
- state the results.
Vague Titles |
Specific Predictive Titles
(Good for Research Proposals) |
Particularly Effective Titles |
| A Study of Aquatic Plants in a Pickle Jar | Elodea Canadensis proposed to have greater [DIRECTION] abundance [DEPENDENT VAR] when in competition for space [INDEPENDENT VAR] with Ceratophyllum demersum in a Model Aquatic Ecosystem [SYSTEM]
|
Addition of caffeine (INDEPENDENT VARIABLE) to aquatic culture in concentrations of 0.1 to 0.5M decreases (DIRECTION) the stem length (DEPENDENT VARIABLE) of Phalaris arundinacea, reed canary grass (STUDY ORGANISM) |
| The Effect of Salt on Aquatic Waterflea, Daphnia magna | Red light expected to increase biomass and average hypocotyl length in Brassica rapa compared to far-red light | *Brine shrimp (Artemia franciscana) grown in acidic water (pH of 3-5) have faster heart rates than brine shrimp grown in water with pH of 7-9 |
*If your report constitutes the results of an experiment where you manipulated variables and analyzed the result, include the independent and dependent variables, the direction of your results, as well as the study organism/ subject in your title.
How will titles be evaluated? To see our expectations for your Title, see the Biocore Research Paper Rubric in this Writing Manual.
Authorship
In scientific journal articles, the first author listed is the primary author, and subsequent authors are listed according to the magnitude of their contribution to the study. Research mentors such as principal investigators (PI’s) of labs, are typically listed last. If all authors have made equivalent contributions to the article, then the paper will state that authors’ names are listed in alphabetical order.
In Biocore you will work within teams to do independent research projects, but we usually ask for individual lab reports because we want to give you many opportunities to work on your writing and thinking skills. At other times we will ask you to submit group posters and PowerPoint presentations. Here is how you should list teammates for various Biocore assignments:
- Individual papers or mini-posters: List yourself first as the primary author under your title, then list teammates as contributors at the top of the page in alphabetical order. Also list your lab section and TA.
- Group posters or PowerPoint presentations: We assume that all of you have made equivalent contributions to these collaborative group assignments, so include all researchers’ names as authors in alphabetical order.
Abstract
*Not all Biocore lab reports require abstracts! Research proposals generally do not require abstracts, but check assignment description for details.
The abstract forces the author to distill the essence of the paper to a very brief summary (100-200 words). Think of the abstract as the two-minute version of your entire experiment. Many readers use the abstract to decide whether they want to find and read the entire paper.
You must be concise. One way to do this is to summarize, in one or two sentences each:
- the rationale behind the experiment (goal of your experiment, model system, most important background information)
- your hypothesis
- the approach you took (how and what you actually tested)
- results or expected results
- conclusions/implications
Other tips:
- Always write the abstract last, after you thoroughly understand the experiment and its meaning.
- Abstracts should be understandable without referring to the rest of the paper.
- You do not cite references in an abstract. General and/or specifically applicable knowledge is assumed or is cited elsewhere in your paper.
Example Abstract From Systematic Observation Study
Adapted from paper by Kristin Magliocco (Fall 2009)
Phosphorus in the runoff to urban streams such as Willow Creek can lead to phosphorus build up and ultimately eutrophication of larger bodies of water. Rain gardens have been constructed on the UW Madison campus adjacent to Willow Creek to prevent accumulation of phosphorus in the creek itself. [Background] By slowing and delaying runoff from reaching the creek, the rain gardens are intended to retain phosphorus and, therefore decrease the amount of phosphorus that reaches the creek. [BR] To test the efficacy of the rain gardens, we hypothesized that there would be no significant difference in the phosphorus concentrations of the water in Willow Creek upstream and downstream of the boundaries of the northeast rain garden. [Hypothesis]We selected four replicate locations in the rain garden itself and in Willow Creek, both upstream and downstream of the rain garden, where we used a Hach phosphorus colorimeter to measure phosphorus concentration. [Approach] Our data supported our hypothesis, with the upstream mean concentration of 0.07335 ± 0.00471 mg/L and the downstream mean concentration of 0.08213 ± 0.0139 mg/L showing no statistically significant difference. [Results]We cautiously concluded that the rain gardens near Willow Creek do prevent further phosphorus accumulation in the stream, but pointed toward future studies focusing on amount of rainfall as an important factor in rain garden efficiency. [Conclusion]
How will abstracts be evaluated? To see our expectations for your Abstract, see the Biocore the Biocore Research Paper Rubric in this Writing Manual.
Introduction
This section provides guidelines on how to construct a solid introduction to a scientific paper including background information, study question, biological rationale, hypothesis, and general approach. If the Introduction is done well, there should be no question in the reader’s mind why and on what basis you have posed a specific hypothesis.
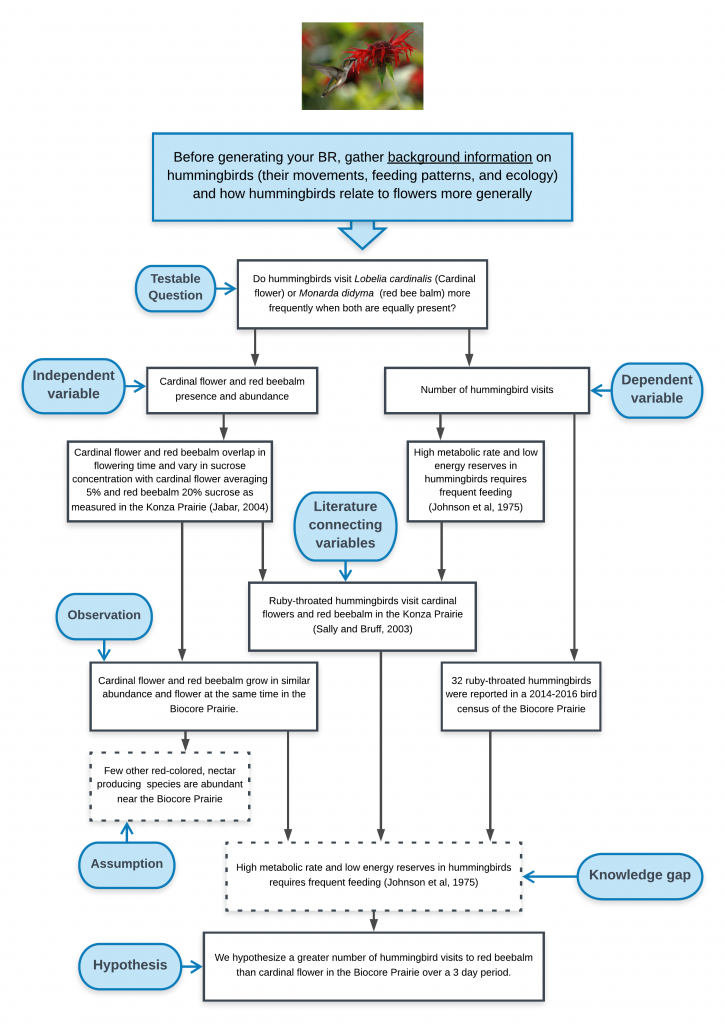
Broad Question: based on an initial observation (e.g., “I see a lot of guppies close to the shore. Do guppies like living in shallow water?”). This observation of the natural world may inspire you to investigate background literature on previous research by others or gather some initial data/ observations as a pilot study. Broad questions are not always included in your written text, but are essential for establishing the direction of your research.
Background information: key issues, concepts, terminology, and definitions are needed to understand the biological rationale for the experiment. The background often includes a summary of findings from previous, relevant studies that introduce the study system, the independent and dependent variable. Remember to cite references, be concise, and only include relevant information given your audience and your experimental design. Your concise summary of background information should lead to specific scientific knowledge gaps that still exist. (e.g., “No studies on lake guppy distribution to date have examined whether guppies do indeed spend more time in shallow water.”)
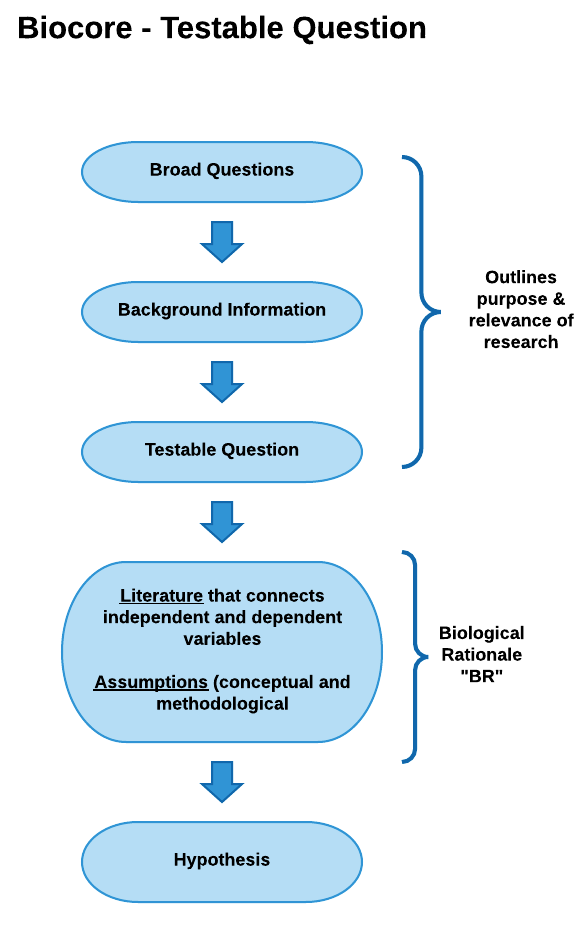
Testable Question: these questions are much more focused than the initial broad question, are specific to the knowledge gap identified, and can be addressed with data. (e.g., “Do guppies spend different amounts of time in water less than 1 meter deep as compared to their time in water that is greater than 1 meter deep?”)
View testable question diagram as pdf
Biological Rationale (BR): The BR explains why you expect your independent variable(s) to affect your dependent variable(s) in the way your hypothesis indicates. After you have summarized the background information relevant to the study, the “BR” provides the logic and reasoning for your hypothesis and experimental approach, describing the biological mechanism that connects your independent and dependent variables and the assumptions that provides evidence for why your hypothesis should be supported. The biological rationale is based on your interpretation of the scientific literature, your personal observations, and the underlying assumptions you are making about how you think the system works. If you have written your biological rationale logically and clearly, your reader should see your hypothesis in your introduction section and say to themselves— “Of course this hypothesis is supportable. It seems very logical based on the rationale presented.”
Steps for Developing a BR—Based on your background information:
- Dependent Variable(s)- List key aspects of the dependent variable (DV) that are known (based on the scientific literature) and those that are unknown that you may need to assume or may be associated with a knowledge gap.
- Independent Variable(s)- List key aspects of the independent variable (IV) that are known (based on the scientific literature) and those that are unknown that you may need to assume or may be associated with a knowledge gap.
- Connection between DV and IV- List what is known and what you are assuming about the ways (mechanisms or relationships) in which the IV influences the DV, either directly or indirectly, either in the system you are studying, in a similar system, or a more distant dissimilar system. If possible, note literature that support any assumptions. The biological link between your IV and DV(s) is central piece of your BR.
- Based on #3, articulate the specific knowledge gap you hope to fill in this study.
- Generate a draft hypothesis based on steps 1-4.
Once you have done steps 1-5, start to sketch out your reasoning using a conceptual or graphic model
In Biocore, we will ask you to construct two different types of models as you are learning to develop your BR:
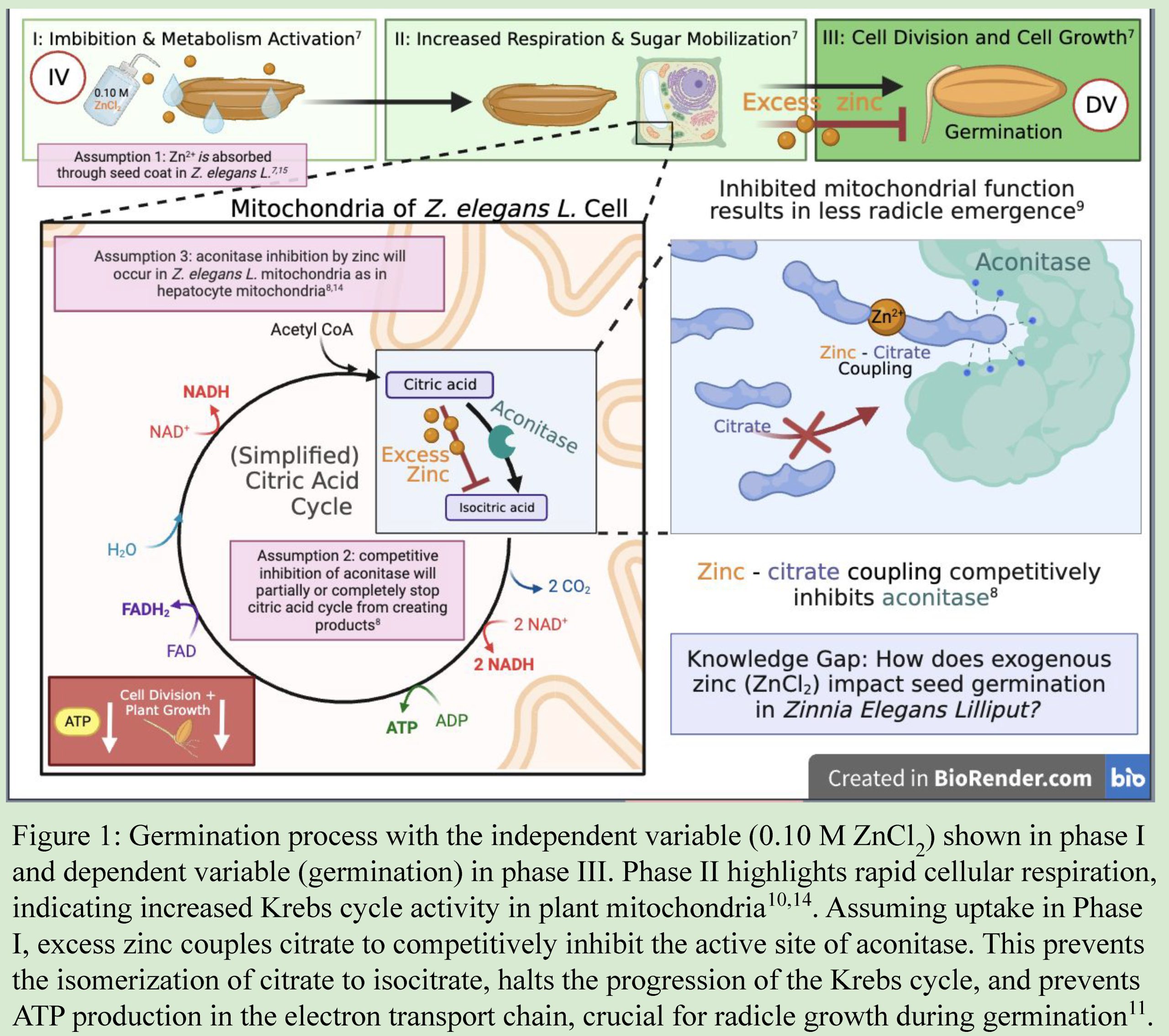
- Conceptual Model– a logical flow of ideas utilizing boxes and arrows to indicate how variables are connected and support your hypothesis. Conceptual models are helpful for developing logical thought progression but are generally not included in a paper or final presentation.
- Graphic or Visual Model– A cartoon or graphic depiction for how variables interact to result in your predicted outcome. Graphic models are often included in scientific posters and Powerpoint presentations, and sometimes in scientific papers.
See following sections for examples of Biological Rationale in the form of Conceptual and Graphic Models
Conceptual Model
In the Conceptual Model example below, the biological rationale is depicted as a logical flow of statements beginning with a testable question and ending with a hypothesis.
View conceptual model as a pdf
Graphic or Visual Model
Graphic or Visual Model uses cartoon diagrams and symbols to communicate the predicted interaction among variables and the mechanism by which they interact. Visual models use shorthand literature citations (superscript numbers) to indicate literature references that are further discussed in an oral presentation (poster or PowerPoint) or written narrative (paper).
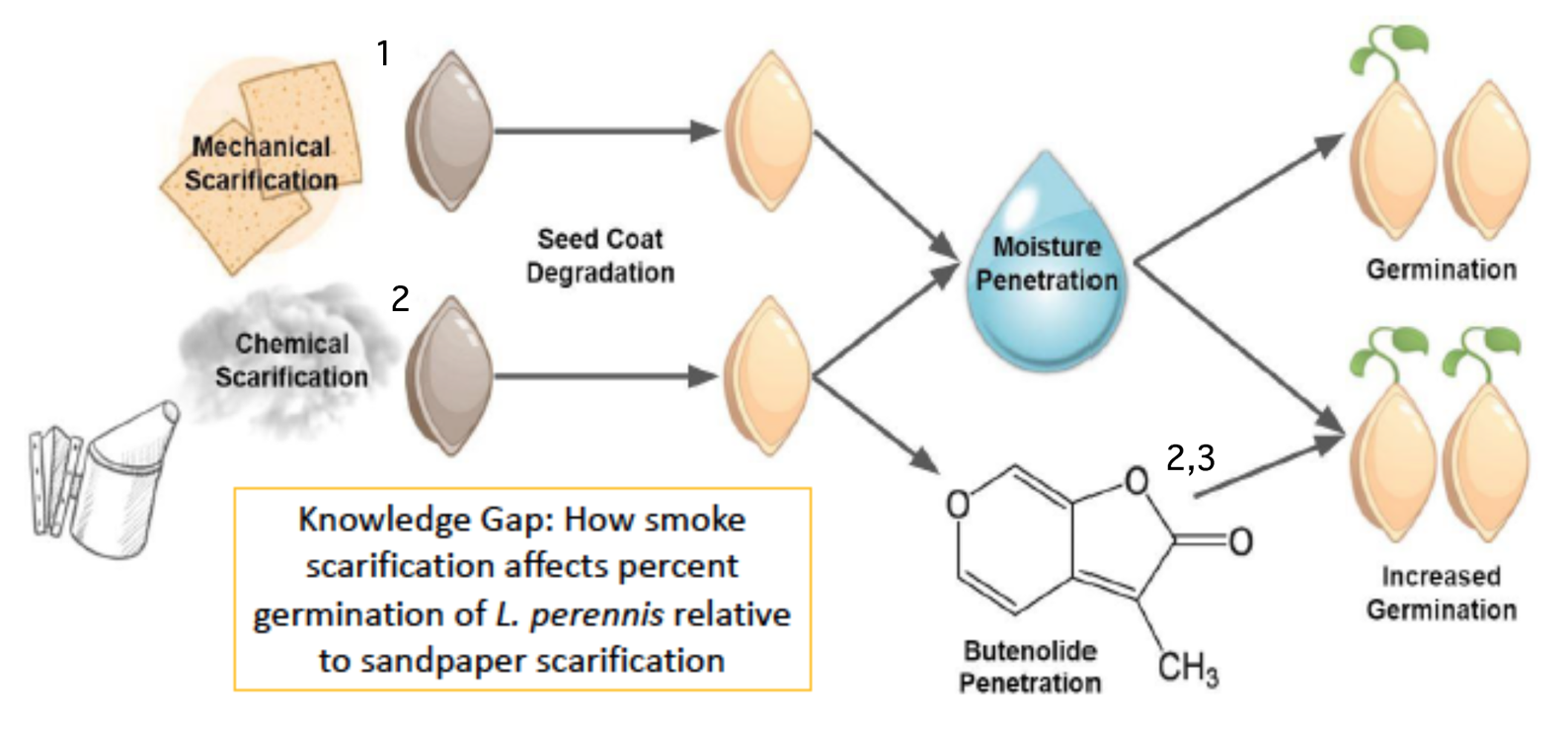
Example Graphic Model of Biological Rationale appropriate for diagram in a paper, poster, or presentation. Adapted from poster by McKenna DeFoer, Sadie Gugel, Evan Polce, Kyrie Sellnow in Biocore 486, Organismal Biology lab.
What do you see done effectively in this example? What would you alter based on the rubric?
Narrative: Scarification using sandpaper abates the seed coat of L. perennis. This process allows moisture to permeate the seed coat during stratification and initiates the biochemical pathway for germination (1. Diboll 2008). Similarly, exposing seeds to cellulose-derived smoke causes chemical scarification (2. Egerton-Warburton 1997). This type of smoke contains butenolide, a compound synthesized during the combustion of plant material that has been found to further stimulate germination (3. Keeley and Fotheringham 1997).
——————————————————————————————————————————————————————–
Example Graphic Model of Biological Rationale appropriate for diagram in a paper, poster, or presentation. Adapted from poster by Lauren Cortright, Trisha Nandakumar, Brian Zheng in Biocore 486, Organismal Biology lab.
What do you see done effectively in this example? What would you alter based on the rubric?
More on Biological Rationale:
- A thorough rationale defines your knowledge gap about the system that has not been revealed in scientific literature or from previous observation. The knowledge gap is the knowledge we are attempting to create. The interpretation of your experimental data and the integration of literature will fill or partially fill the knowledge gap. In order to fill the knowledge gap, you may need to make assumptions about how your system operates. Assumptions are aspects of the system that you are not testing directly, but you think are particularly important since they drive the direction of your specific hypothesis or general predictions. Sometimes students confuse the knowledge gap and assumptions. Data gathered during the experiment can address the knowledge gap but generally do not provide direct evidence to support or refute assumptions.
- Defining the BR is probably the most critical task for a writer, as it tells your reader why your research is biologically meaningful. It may help to think about the rationale as a link between your independent and dependent variables, because the rationale answers these questions—how is this investigation related to what we know, what assumptions am I making about what we don’t yet know, AND how will this experiment add to our knowledge?
- Expect to spend time and mental effort on your BR. You may have to do considerable digging into the scientific literature to define how your experiment fits into what is already known and why it is relevant to pursue.
- Be open to the possibility that as you work with and think about your data, you may develop a deeper, more accurate understanding of the experimental system. You may find the original rationale needs to be revised to reflect your new, more sophisticated understanding.
- As you progress through Biocore and upper level biology courses, your rationale should become more focused and matched with the level of study i.e., cellular, biochemical, or physiological mechanisms that underlie the rationale. Achieving this type of understanding takes effort, but it will lead to better communication of your science.
Hypothesis / Predictions: specific prediction(s) that you will test during your experiment. For manipulative experiments, the hypothesis should include the independent variable (what you manipulate), the dependent variable(s) (what you measure), the organism or system, the direction of your results, and comparison to be made. See the following examples.
| Hypothesis that Needs Work (manipulative experiment) |
Better Hypothesis (manipulative experiment) |
| We hypothesized that Daphnia magna reared in warm water will have a greater sexual mating response.
[The dependent variable “sexual response” has not been defined enough to be able to make this hypothesis testable or falsifiable. In addition, no comparison has been specified— greater sexual mating response as compared to what?] |
We hypothesized that Daphnia magna (STUDY ORGANISM) reared in warm water temperatures ranging from 25-28 °C (IND. VAR.) would produce greater (direction) numbers of male offspring and females carrying haploid egg sacs (DEPEND. VAR.) than D. magna reared in cooler water temperatures of 18-22°C. |
If you are doing a systematic observation, your hypothesis presents a variable or set of variables that you predict are important for helping you characterize the system as a whole, or predict differences between components/areas of the system that help you explain how the system functions or changes over time.
| Hypothesis that Needs Work (systematic observation) |
Better Hypothesis (systematic observation) |
| We hypothesize that the frequency and extent of algal blooms in Lake Mendota over the last 10 years causes fish kills and imposes a human health risk.
[The variables “frequency and extent of algal blooms”, “fish kills” and “human health risk” have not been defined enough to be able to make this hypothesis testable or falsifiable. How do you measure algal blooms? Although implied, hypothesis should express predicted direction of expected results (e.g. higher frequency associated with greater kills). Note that cause and effect cannot be implied without a controlled, manipulative experiment.] |
We hypothesize that increasing (DIRECTION) cell densities of algae (VAR.) in Lake Mendota over the last 10 years is correlated with 1. increased numbers of dead fish (VAR.) washed up on Madison beaches and 2. increased numbers of reported hospital/clinical visits (VAR.) following full-body exposure to lake water. |
Note that hypotheses/ predictions you develop in Biocore lab are much more specific than the general hypotheses that guide the research questions you encounter in scientific literature or in faculty research labs. That is because the research projects you do in Biocore are short-term, small(er) in scale or context specific, and therefore require greater specification to be testable within our class context.
Experimental Approach: Briefly gives the reader a general sense of the experiment, the type of data it will yield, and the kind of conclusions you expect to obtain from the data. Do not confuse the experimental approach with the experimental protocol. The experimental protocol consists of the detailed step-by-step procedures and techniques used during the experiment that are to be reported in the Methods and Materials section.
***Some Final Tips on Writing an Introduction***
- As you progress through the Biocore sequence for instance, from organismal level of Biocore 381/382 to the cellular level in Biocore 383/384, we expect the contents of your “Introduction” paragraphs to reflect the level of your coursework and previous writing experience. For example, in Biocore 384 (Cell Biology Lab) biological rationale should draw upon assumptions we are making about cellular and biochemical processes.
- Be Concise yet Specific: Remember to be concise and only include relevant information given your audience and your experimental design. As you write, keep asking, “Is this necessary information or is this irrelevant detail?” For example, if you are writing a paper claiming that a certain compound is a competitive inhibitor to the enzyme alkaline phosphatase and acts by binding to the active site, you need to explain (briefly) Michaelis-Menton kinetics and the meaning and significance of Km and Vmax. This explanation is not necessary if you are reporting the dependence of enzyme activity on pH because you do not need to measure Km and Vmax to get an estimate of enzyme activity.
- Another example: if you are writing a paper reporting an increase in water flea heart rate upon exposure to caffeine you need not describe the reproductive cycle of water fleas unless it is germane to your results and discussion. Be specific and concrete, especially when making introductory or summary statements.
Where do you discuss Pilot Studies?
Many times it is important to do pilot studies to help you get familiar with your experimental system or to improve your experimental design. If your pilot study influences your biological rationale or hypothesis, you need to describe it in your Introduction. If your pilot study simply informs the logistics or techniques, but does not influence your rationale, then the description of your pilot study belongs in the Materials and Methods section.
How will introductions be evaluated? To see our expectations for your Introduction, see the Biocore Research Paper Rubric in this Writing Manual.
Example Introductions
Example Introduction From Systematic Observation Study
Adapted from a paper by Will Klein 2009
Throughout history, humans have discovered and used chemicals derived from plant extracts as antimicrobial compounds for medicinal purposes. Although useful to humans, why would a plant create an antimicrobial defense that affects the growth of bacteria? [broad study question] As non-mobile organisms, plants have evolved mutually beneficial associations with beneficial microbes (Brooker et al. 2011) and a full arsenal of adaptations for defense against pathogenic microorganisms (bacteria, viruses, fungi). Borchardt et al. (2008) did an antimicrobial screening of 339 plant species growing in Minnesota and Wisconsin, many of which are prairie plant species. The researchers tested aerial plant parts (leaves, stems, flowers) for growth inhibition of one, two or three common mammalian pathogens (Escherichia coli, Staphylococcus aureus, Candida albicans) and found 109 species inhibited growth of at least one microorganism. Leave extracts of Silphium sp., a species found in the Biocore Prairie, contains antimicrobial compounds that inhibit the growth of many types of Gram-negative and Gram-positive bacteria (Kowalski and Kedzia, 2007; Kowalski, 2008). [background information]
Plants may produce chemical defense in the form of antimicrobial compounds contained in stems, roots, leaves, bark, flowers or fruits. [BR: assumption] By investing energy to generate these antimicrobial compounds, the plant maximizes its likelihood to succeed in its particular ecological niche (i.e. the Biocore Prairie) and improves its biological fitness. [BR: assumption] No studies howver have directly examined the effect of native Biocore prairie plant extracts on indigenous soil bacteria growth. [testable question]
Through preliminary investigations in the Biocore Prairie during summer 2010, we sought to find prairie plant species and extracts from different plant parts (roots, leaves or stems) that would inhibit soil bacteria-bacteria cultured from soil that the prairie plants are growing in. Although most soil bacteria are beneficial or do nothing to affect prairie plants, we reasoned that plant species coexisting in the same environment with particular soil microbes may have efficient defense mechanisms towards pathogenic “prairie soil” bacteria. [BR: assumption] Huechera richardsonii, Monarda fistulosa, and Euphorbia corollata are three species common to the Biocore Prairie. Although leaf tissue of these three species have all been shown to contain antimicrobial properties against S. aureus (Borchart et al. 2008), how extracts from these species influence growth of bacteria indigenous to the Biocore Prairie is not known. [knowledge gap] We believe these plant species will contain antimicrobial properties in leaves to protect the tissue from microbial leaf pathogens that also occur in the soil. [BR: assumption]
We hypothesized that leaf extracts of Huechera richardsonii, Monarda fistulosa, and Euphorbia corollata would exhibit antimicrobial properties on the bacteria found in their native environment. [hypothesis] Our approach was to grow soil bacteria collected from the Biocore Prairie on agar plates, and then expose bacteria to leaf extracts absorbed on filter paper discs and measure the extent to which the extracts inhibited bacterial growth. [approach]
*Note: If you are a Biocore 382 student—do not worry if you don’t understand the scientific content in these two examples. We will get there! These examples are provided to refer to as you progress through the curriculum
Example 4: Good Introduction from manipulative experiment in Cell Biology Lab
(adapted from a poster by Kari Esselman, John Kinzfogl, Amber Kugel, & Katie Luettgen, Spring 2003)
In the yeast (Saccharomyces cerevisiae) mating signal transduction pathway, interaction of the complete –mating factor with
the G-protein-coupled receptor on a MAT-a cell induces cell cycle arrest in the G1 phase, morphological changes or “shmooing,” and activation of genes involved in the mating process (Hoopes et al., 1998). In Saccharomyces cerevisiae, the amino acids Trp1, Lys7 and Gln 10, the central ß –turn conformation, and the amino acids near the C-terminus are directly involved in the binding of the a–mating factor to the receptor (Saskiawan et al., 2002). Altering the structure of the a –factor produces a conformational change in the receptor that is distinct from the conformational change of the normal a –factor, consequentially altering or even inhibiting the mating cascade of events (Bukusoglu and Kemmess. 1996). Elimination of Lys7 and Gln10 from the a –mating factor results in greater than a 100 fold decrease in mating signal transduction (Xue et al., 1996). [all background info]
It is unclear whether elimination of amino acid residues other than Lys7 and Gln10 in the a –mating factor also decrease the yeast mating response. [broad question] When introduced to MAT-a Saccharomyces cerevisiae cells, this sort of a – factor fragment could: 1.bind to the receptor site and induce the same change that the complete a –mating factor would; 2. bind to the receptor site but not induce the same changes as the complete a –factor, or 3.not bind to the receptor site at all. [BR: assumed biological mechanism] If the mating response to this fragment is different than normal (BR: assumption), this would indicate which amino acid side groups are important in binding the receptor. An examination of Saccharomyces cerevisiae response to an a –mating factor fragment missing amino acids other than Lys7 and Gln10 would thus increase our understanding of the specificity of the a –factor receptor for its ligand. [BR: study goal/broader implication]
We hypothesized that the introduction of an a –mating factor fragment missing amino acids 7 through 13 to MAT-a Saccharomyces cerevisiae cells would cause more budding and less mating gene transcription and shmooing, as compared to the response to the complete a –factor. [hypothesis] We tested this hypothesis by adding this a –factor fragment to yeast cells transformed with a plasmid containing the FUS1 promoter attached to the lacZ reporter gene and recording the resulting morphological changes (budding and shmooing) and ß-galactosidase (ß -gal) activity. [approach]
Example 5: Good Introduction from manipulative experiment in Organismal Biology Lab
(adapted from a paper by Matt Young, Fall 2003)
The diving response is a set of characteristic reactions following the immersion of certain body parts in water. It is observed primarily in diving mammals and ducks, but humans have also elicited the response, perhaps as a trait that was not selected against during their evolution (McCulloch et. al. 1995; Hlastala and Berger 2001). Gooden (1993) clearly demonstrated that the diving reflex prepares the animal’s body for the effects of long periods of apnea (breathing cessation) associated with being underwater. It does this by decreasing oxygen consumption and redirecting blood flow out of the peripheral structures and towards the central organs such as the heart and brain.
McCulloch et. al. (1995) showed that the diving response is initiated by the stimulation of the trigeminal (Vth cranial) nerve, a primary sensory supply from the face, including the nose and forehead areas. Stimulation of this nerve results in a complex series of sympathetic and parasympathetic nerve activations (Gooden 1994). Increased parasympathetic activity triggers the vagus nerve to inhibit the cardiac pacemaker, resulting in reduced heart rate (Andersson et. al. 2000). Limb vasoconstriction occurs in response to increased sympathetic nerve activity, which results in increased mean arterial blood pressure (MABP) (Andersson et. al. 2000; Gooden 1994). [all background info in previous paragraphs]
Along with submersion in water, apnea is believed to be a major component in eliciting a proper diving response. It is still not clear, however, how necessary apnea is for the induction of the diving response or the mechanism for this induction (Gooden 1994). [broad question] Campbell et. al. (1969) argued that apnea, whether voluntary or involuntary, is essential for a diving response to occur, while Andersson et. al. (2000) found that facial immersion with eupnea resulted in reduced, but noticeable, diving responses. [background info]
Along with submersion in water, apnea is believed to be a major component in eliciting a proper diving response. It is still not clear, however, how necessary apnea is for the induction of the diving response or the mechanism for this induction (Gooden 1994). [broad question] Campbell et. al. (1969) argued that apnea, whether voluntary or involuntary, is essential for a diving response to occur, while Andersson et. al. (2000) found that facial immersion with eupnea resulted in reduced, but noticeable, diving responses. [background info]
It is believed that apnea stimulates chemoreceptors and thoracic stretch receptors in order to exert its effects. The thoracic stretch receptors are sensitive to movements in the airways, while chemoreceptors are sensitive to the oxygen lack associated with breath-holding. Increased firing of these two receptors due to their respective stimuli is believed to be the method by which apnea influences the diving response, but the exact pathway this firing takes to exert such effects remains unclear. It may either directly affect the cardiovascular centers, or indirectly affect the cardiovascular system via the medulla (Gooden 1994). [background info which identifies knowledge gap]
Does apnea significantly increase the human diving response during facial submersion? [testable question] It seems plausible that simultaneous activation of the trigeminal nerve, thoracic stretch receptors, and arterial chemorecptors would produce a more pronounced cardiovascular diving response.(BR: biological assumption) The goal of this experiment is to examine whether the diving response in eupneic (normal breathing) situations is significantly different than that observed during apneic situations. [BR: study goal]We will focus on heart rate and blood pressure changes, two of the many responses associated with the diving response. If heart rate and blood pressure changes during apneic submersion are significantly greater than those observed during eupneic submersions, this would indicate that simultaneous stimulation of the trigeminal nerve, thoracic stretch receptors, and chemoreceptors produces a greater cardiovascular response than stimulation of the trigeminal nerve alone. [BR: assumed mechanism]
We hypothesized that diving responses in human participants would be more pronounced in those experiencing apnea during immersion compared to those experiencing eupnea. More specifically, we expected non-breathing participants’ heart rates to decrease and blood pressures to increase significantly more than breathing participants in response to facial immersion in cold water. [hypothesis]
We tested this hypothesis by having 12 human subjects immerse their foreheads, noses, and cheekbones in cold water. We used a paired analysis to determine whether the change in heart rate and blood pressure from just prior to immersion to the end of immersion was different during apneic as compared to eupneic submersions. [approach]
Methods and Materials
This section is often the easiest to write since it is simply a clear explanation of the specific procedures, techniques, and materials you used. In some cases (e.g., the projects carried out in the Biocore Prairie), it is necessary to include procedures carried out by previous classes as well. Provide enough details that a knowledgeable reader (e.g., a Biocore peer who is not enrolled in lab) could replicate the experiment. This will also allow him/her to evaluate whether to trust your findings. In the case of field investigations, include a description of the type of community and the location of the site studied.
Mathematical manipulations or statistical analyses applied to the data should be explained under a subheading, but keep these brief. Although calculations are not normally included in a scientific paper, we sometimes ask you to include examples to check whether you are doing them correctly. If this is the case, put them in an appendix at the end of the paper.
Focus on essentials that affect the results. For example, in a genetics experiment with flies, it is important to state whether the females used for the crosses were virgins; it is not necessary to list the type of food or anesthetic used. However, these details would be important if your experiment was testing how different diets affected fruit fly activity level or some other physiological parameter. In cases where detailed protocols are given in the lab manual, merely cite the appropriate chapter of the lab manual, note any details relevant to the experiment but not specified in the protocol (e.g., identify the particular strain of organism you and your teammates used when several were available), and describe any manipulations you made that are not outlined in the manual. Include only what is vital for the reader’s understanding of how the results were obtained. (E.g., Drawing white poker chips out of a 1 quart Babcock Vanilla flavored ice cream container to get two numbers to pace out and place quadrats is not as important as the fact that quadrat placement was random.) If you are having trouble deciding what to put in and what to leave out, consult with your TA, peers, or other instructional staff for guidance before handing in your final paper.
 Organize the procedures in the Methods & Materials section logically:
Organize the procedures in the Methods & Materials section logically:
- Use subheadings, including one called “data analysis”
- Describe your schedule of procedures in chronological order (if it makes sense to do so)
- When writing a final paper, use the past tense for this section (because you refer to procedures that you carried out in the past). When writing a proposal, use future tense.
- Report final concentrations (in molar, millimolar, micromolar etc). rather than final volumes (see table below). Readers can replicate concentrations, but often find it difficult to discern concentrations when only volumes are reported.
| Not helpful to other researchers | Very helpful |
| Reporting final volumes. E.g., ‘We added 5 ml of NaCl solution to the reaction mixture.’ | Reporting final concentrations. E.g., ‘The final reaction mixture contained 2 mM of NaCl.’ |
Example of Good Methods text
(Excerpt adapted from a paper by Beth Theusch, Biocore 384, Spring 2003: Inorganic phosphate competitively inhibits alkaline phosphatase-catalyzed hydrolysis of p-nitrophenylphophate)
Pilot Study*
A pilot study using various Pi concentrations but a constant substrate concentration close to the Km value was conducted in order to determine a Na2HPO4 concentration that has a moderate effect on initial reaction velocity to use in the inhibitor kinetics study. We tested a range of concentrations between 2.5 uM and 200 uM Na2HPO4 in tubes containing 0.05 M Tris-HCl, pH 8.6, 0.05 mM pNPP (the approximate Km value), and 4 ug/ml bovine intestinal alkaline phosphatase in a total volume of 5 ml. There was a control with no Na2HPO4 added and a blank with no enzyme added.
Experimental Protocol
The inhibitor kinetics study involved two sets of replicated reactions over a 0-0.5 mM range of pNPP substrate concentrations. One set of reactions was conducted in the absence of inhibitor and used as a control. The other set of reactions had a uniform concentration of Pi inhibitor, which was determined to be 0.05 mM from the pilot study, added to each tube. All tubes had 0.05 M Tris pH 8.6, 4 ug/ml alkaline phosphatase, and the appropriate amount of distilled water to bring the total volume of each tube to 5 ml. In each case, there was a control with no substrate added and a blank with no enzyme added. The pH of the Na2HPO4 salt solution was checked to ensure that the pH was approximately the same in the uninhibited and the inhibited reactions. Four replicates were performed for both the inhibited reaction and non-inhibited reaction.
For a complete protocol of the non-inhibited experiment, refer to “Enzyme Catalysis” in the Biocore Cellular Biology Lab Manual (Becker, Metzenberg, Dehring, 2003). For the inhibitor kinetics study, the product concentrations were used to calculate the initial reaction velocities at each substrate concentration in the presence and absence of inhibitor. Michaelis-Menten curves and Lineweaver-Burk plots were then generated to compare the values of Km and Vmax for the inhibited and uninhibited reactions. Ki was determined using the relationship that the inhibited Km = (1 + [inhibitor] / Ki) times the uninhibited Km.
Statistical Analysis
We performed an independent sample T-test to determine whether the differences between the average Km and Vmax values between the inhibited and uninhibited reactions were statistically significant.
*Note: Not all papers require the inclusion of pilot studies in the Methods section. Discuss this with your instructors.
How will methods/materials be evaluated? To see our expectations for your Methods & Materials, see the Biocore Research Paper Rubric in this Writing Manual.
Results
The Results section is a logically organized presentation of your observational and numeric data. This is an opportunity to emphasize points or trends that you will be focusing on in your discussion. In many cases the organization and subheadings of this section should be consistent with those of the Methods and Materials section.
Before you start writing, make sure you have discussed the data and have shared your plan for analysis with your group members. Your group should share a common data set and, therefore, should be working with the same mean, standard deviation, and other descriptive statistics. As long as all group members have the same raw data set, you may choose to display the data differently.
There are usually two parts to this section:
- text
- tables and figures
Text: The key purpose of the text in the results section is to point out and emphasize patterns in your data. You may choose to illustrate some of these patterns, especially those that pertain to your hypothesis, in figures or tables. However, each figure and table needs accompanying text to point out the obvious—or sometimes the not so obvious.
- Briefly describe, but do NOT make conclusions about (i.e., interpret) your data here—save that for the Discussion section.
- Point out any trends. (Trends are relationships between one variable and another. e.g., as variable one changes, variable 2 tends to change in a consistent way.)
- Note differences or similarities between treatment groups.
- If you perform statistical analyses, report any significant biological differences you found, followed by pertinent statistical summary information (test score such as a “t” or “F” value, degrees of freedom, one or two-tailed p-value; see Biocore Statistics Primer for more info).
| Not helpful to other researchers | Very helpful |
| We found statistical significance. | The average reaction velocity (54 nmol/min, SE ±0.084) in the inhibited reactions was lower than the average uninhibited reaction velocity (25 nmol/min, SE ± 0.12). This difference was statistically significant (t(12)=135.4, p<0.001, two tailed). |
Refer your reader to “Table 1” or “Figure 1” as you explicitly identify relationships, patterns, or general trends that you see in the data. Remember that relationships that are obvious to you may not be obvious to someone who has not carried out the experiment.
- Never write a sentence that just tells the reader where the data are. Point out to your reader the general trends in the data, then refer to the figure or table parenthetically.
- When using the term “significant” in your results section recognize that it has a specific connotation in science that reads “statistically significant.” Therefore, use the term “significant” when explaining differences you observe only if you found statistically significant differences.
The Results section should not be controversial since you are merely reporting findings, not saying what you think they mean. Avoid judging your data as “good” or “bad.” Data are facts and facts simply are what they are. Remember: you are not graded on whether your experiment “worked” or on your results; you are graded on how you handle them. Always report what you saw, not what you think you should have seen.
See the following excerpt from a good Results section describing data from a systematic study.
Example of a Good Results Section from a Systematic Observation Study
(excerpted from a Biocore 382 paper by Kim Treml, Fall 2003)
Results
Water Quality
Water quality testing revealed a mean pH of 6.67 +/-0.07 pH units (Table 1). Mean dissolved oxygen and dissolved carbon dioxide were 3.4 +/- 0.4ppm and 55 +/-3ppm respectively. Also, the total phosphorus was measured as 0.51 +/-0.5mg.L and conductivity, measured in microsiemens, was 1,063 +/-17μs. All means were computed with n=45. Both conductivity and phosphorus fall far out of range of optimal water quality levels for a healthy aquatic ecosystem (Table 1). The measured phosphorus level is an order of magnitude larger than what is recommended by the EPA. Conductivity is twice as high as the ideal level in a freshwater ecosystem. [RESULTS TEXT]
Table 1. Water quality data obtained from the University Bay marsh in 2003. Each value represents the mean of 45 trials. The error margin is + or – 1 standard error. Optimal data ranges for a healthy aquatic ecosystem are shown for comparison. [TABLE LEGEND]
| Parameter | 2003 data | Optimal data ranges |
| pH | 6.57 +/- 0.07 | 6.9 – 7.1 |
| Conductivity (μs) | 1,063 +/ 17 | 150 – 500 |
| Dissolved O2 (ppm) | 3.4 +/ 0.4 | 5 – 6 |
| Dissolved CO2 (ppm) | 55 +/ 3 | > 20 |
| Phosphorus (mg/L) | 0.51 +/ 0.04 | 0.005 – 0.05 |
[TABLE]
Macroinvertebrate Diversity
Macroinvertebrate species in the University Bay marsh were catalogued and presence or absence of each species was noted. Figure 3* depicts the calculated frequency of each species per 500mL. The species are approximately organized on the chart from left to right with increasing pollution tolerance as described on North Carolina State University’s water quality webpage (2003). The highest frequency in both 2002 and 2003 exists among organisms around the mid-range of pollution tolerance. Orb snails, scuds, backswimmers, copepods, seed and clam shrimp, nematodes and tubifex worms were present in over half of our samples in either 2002 or 2003. Species indicative of very high water quality or very low water quality were less frequent compared to species indicative of the mid range. Nonetheless, the data show an increase in the variety of species present from 18 species in 2002 to 26 in 2003. [RESULTS TEXT]
*Figure 3 not shown in this Writing Manual
Tables and Figures:
Tables and figures are key elements of a scientific paper.
- Tables are organized lists of numbers, ideas, or other data.
- Figures are graphs, charts, diagrams, or photos.
Why use tables and figures? First, they offer a concise way to present a large amount of information. Second, they carry the bulk of the experimental evidence needed to support your conclusions. Third, they offer the reader a chance to assess your data and determine whether or not your conclusions are valid. Finally, the values in them can be used by other scientists who wish to build on your work. Usually, summarized (e.g., averages and measures of variation) rather than raw data are included in a paper. Always make it clear whether you are presenting actual data or averages. (In some cases we will ask you to include raw data as an appendix.) Please refer to the Biocore Statistics Primer for directions on producing figures in Excel.
Each table or figure should be referred to in the text of your paper at least once. If you have nothing to note about a particular table or figure, leave it out. Identify and number tables or figures according to the order they appear in the text (Table 1, Table 2, Figure 1, Figure 2, etc.). This way the reader will know exactly what data you are discussing.
Tables and figures should be neat, logically organized, and informative. If properly prepared, they can stand independently of the paper. Always remember that readers are not familiar with your data. A table or figure that seems self-explanatory to you may not seem so to a reader.
Here are some rules for presentation of graphs and tables:
- Present your final data in table or graphical form. The choice of table or figure should be based on the type of data you have. If you are trying to show trends or simple comparisons it may be best to use a figure. If you have long lists or many comparisons to be made across groups a table may be more appropriate. [DO NOT present the same data in both table and graphic form.]
- The most common way to present graphical data is either an XY scatterplot for continuous data or a bar chart for categorical data/ results of statistical comparison of the means of two or more groups.
- Keep it simple! The amount of time it takes a reader to interpret a figure is inversely proportional to how well those data are presented. Do not overuse transformations or ratios if they are unnecessary for accuracy and clarity of your results.
- Clearly label all axes or columns including units (e.g. Time (min.), Concentration (mM), Mass (mg)). Describe any symbols you use in your graphs using a KEY (see figures below for examples of keys).
- Table and figures should always have a brief text description called a LEGEND that fully describes them so that they can stand alone. (See figures below for examples of figure legends.)
- POOR LEGEND: Enzyme activity vs. salt (Avoid using the term vs)
- BETTER: Average alkaline phosphatase activity for concentrations of NaCl from 0.1 to 1 mM. The substrate for the reaction was ATP at a concentration of 2 mM for a total reaction time of 3 minutes. Columns represent mean values (N=3) with error bars representing ± 1 SE.
- Put table legends above a table. Put figure legends below or to the side of a figure.
- Do NOT create titles for figures or tables. Instead of a title, use a simple legend numbering each table and figure consecutively is sufficient. Do not use titles like “Chart 1” that are automatically generated by Excel.
- For graphs that present an average value as a single point or bar, include error bars and state what they represent. Usually, this will be 1 standard deviation (SD) or 1 standard error (SE) on either side of the mean (see figure 1 below for an example).
- For tables presenting means, include some measure of variation (SD or SE). (See Table 1 above for an example of this).
- State the number of samples used to calculate an average. If you measured the height of 12 purple cone flower plants and reported an average height of 0.82m, indicate the number of samples used to generate that statistic as n=12.
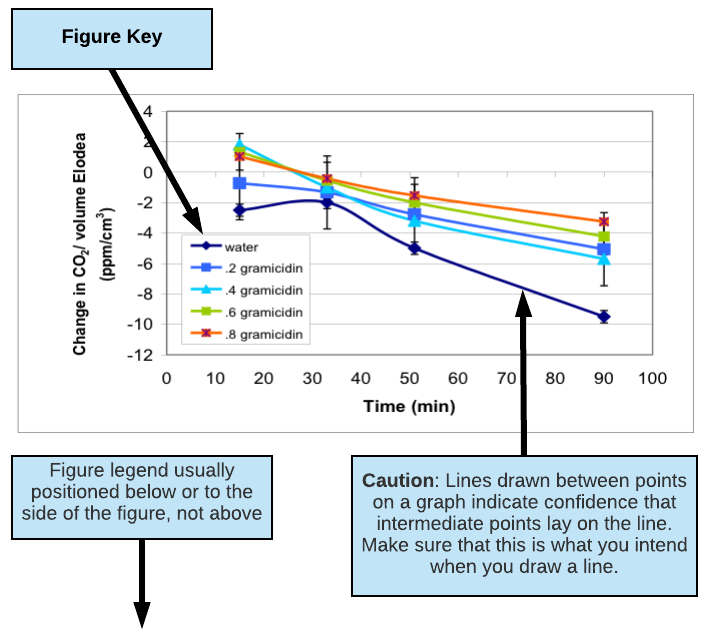
- Do not connect the points on a line graph unless you really mean to say that the values in between the points shown should follow the line drawn. Trend lines have very limited predictive value or validity when connecting 3 points or less.
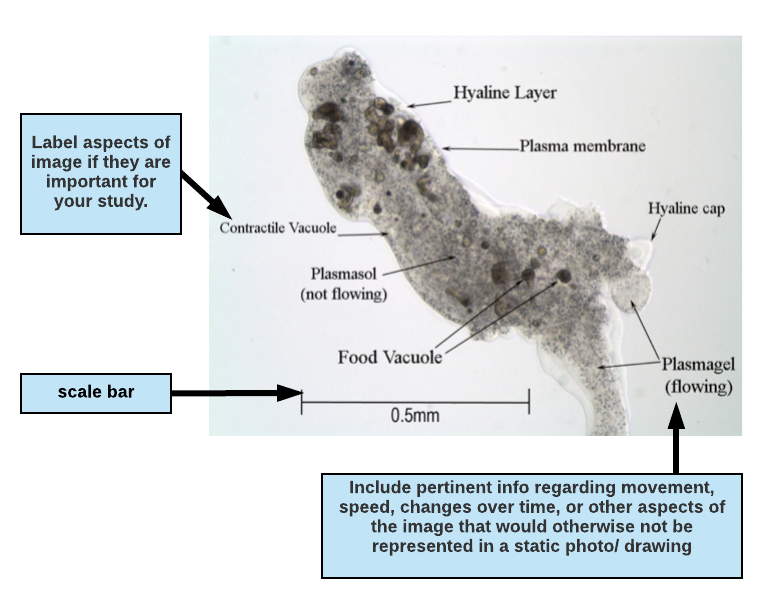
Drawing a diagram or presenting a photomicrograph: Drawn diagrams or photographs taken from a microscope and their legends should contain enough information that a reader can understand (as near as possible) what you actually observed and the conditions surrounding the observation. Diagrams must be large enough to show significant details of what you observed. In practice, this generally means that each diagram should cover at least a quarter of an 8.5×11” page. Indicate the type of microscopy used and the total magnification in your legend. Include a scale on your drawing. Define the experimental conditions and include notes on the process of your investigation. See Figures A-7, A-13, and A-14 in the World of the Cell’s “Principles & Techniques of Microscopy” for examples of good figure legends.
Example of Good Results bar graph
(excerpted and adapted from a paper by Jo Ellen Lomax, Biocore 486, Fall 2003; Speed of Chloroplast Movement) 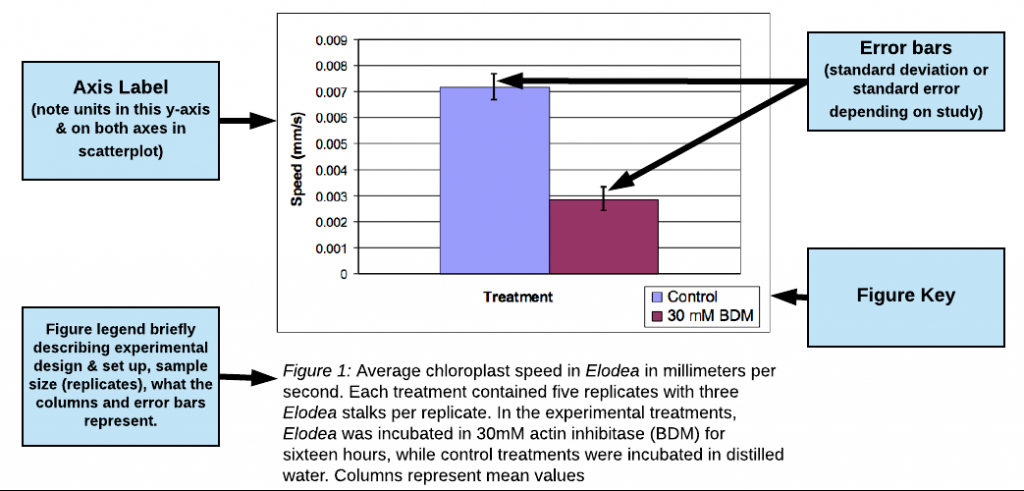
view this figure as a pdf
Example of Good Results scatterplot
(excerpted and adapted from a presentation by Jennifer Rowland, Beth Rollmann, Simona Rosu, and Christopher Luty, Biocore 384, Spring 2003; Gramicidin Decreases CO2 Consumption in Elodea)
Figure 2: Change in dissolved CO2 levels in water surrounding six Elodea sprigs (6 cm in length) in 75 ml culture tubes over 100 minutes of light exposure. Dissolved gramicidin concentrations ranged from 0 to 0.8 µM. Each data point represents the mean of N=11-15 culture tubes for each gramicidin concentration plus/minus one standard error.
Example of Good Table
(adapted from Jenna Voegele paper on water quality in Willow Creek, Biocore 382, Fall 2004)
Table 1. Mean values of water chemistry tests from upstream and downstream sampling locations during a three day study period, Sept 14-16, 2004. Variation is shown as ± 1 SE next to each mean value, followed by sample size (in parentheses) in which varied for each test and sampling location. Note the smaller sample size for the nitrate-N test.
| Sampling Location
|
||
| Water Quality Test | Downstream | Upstream |
| Turbidity (NTU) | 32.2 ± 9.7 (16) | 23.6 ± 5.9 (13) |
| PH | 6.99 ± 0.1 (16) | 6.97 ± 0.12 (14) |
| Dissolved Oxygen Saturation (%) | 77.1 ± 1.7 (32) | 81.5 ± 1.9 (26) |
| Biochemical Oxygen Demand (mg/L) | 2.6 ± 0.5 (20) | 3.3 ± 0.7 (18) |
| Total Phosphorus (mg/L) | 0.44 ± 0.09 (15) | 0.58 ± 0.12 (14) |
| Nitrate-N (mg/L) | 8.6 ± 1.4 (4) | 11.0 ± 0.7 (4) |
| Water Temperature (°C) | 20.8 ± 0.3 (17) | 20.6 ± 0.3 (14) |
| Fecal Coliform (colonies/100ml water) | 414 ± 185 (29) | 684 ± 201 (24) |
Writing a figure legend for a drawing or micrograph:
If you are including an image (drawing or photomicrograph) in your paper, highlight attributes of the image that are important for your paper and to your reader. If the reason for including the image is to highlight anatomy, you may want to label structures and include a description of movement or other important observations in the figure legend. When writing a figure legend to accompany a photo or drawing, include enough information so that a reader can understand (as near as possible) what you actually observed and the conditions surrounding the observation. This means that you should indicate the type of microscopy used (phase contrast, bright field, fluorescence, etc.) and any notes regarding the preparation (e.g., mounted in ProtoSlow, water or saliva, with coverslip, types of stains used, etc.). Also indicate the total magnification in your legend. Diagrams must be large enough to show significant details of what you observed. It is important to include a scale on your drawing.
Click on the three purple icons in the diagram below for more information about each element.
Figure 1.1 Micrograph of the protozoan Pelomyxa carolinensis viewed under phase contrast microscopy, magnification 100X. The specimen is mounted in ProtoSlow and coverslip to reduce its movement. Plasmagel streams readily into pseudopodia (seen at the bottom right of the photo) allowing the amoeba to slowly crawl across the field of view.
In the figure descripton above, the writer has indicated the type of microscopy (phase contrast microscopy, magnification 100X) and the total magnification (100X).
How will results (including text & figures/tables) be evaluated? To see our expectations for your Results, see the Biocore Research Paper Rubric in this Writing Manual.
Discussion
This is where you interpret your results for the reader. It is the most important part of your paper and often one of the most difficult to write. The discussion section is NOT a restatement of your results, but rather where you provide your insight on the investigation through logical analysis. Key elements of your discussion section include:
- BROAD STUDY QUESTION that your research is trying to address
- SUPPORT/REJECT HYPOTHESIS
- INTERPRET the dependent variable measured (if multiple variables are measured you interpret each variable independently and then INTEGRATE variables for overall interpretation of data)
- Formulate argument for your conclusions, emphasizing how your data do or do not support your biological rationale & by comparing with relevant findings in the literature
- NEW KNOWLEDGE that your investigation has generated: highlight the knowledge gap that your data help address, and the implications of your work. Introduce at least one new paper from the scientific literature to help you discuss or support your findings.
- EVALUATE confidence in experimental design and reliability of data
- NEW QUESTIONS and FUTURE STUDIES that the new knowledge inspires
- UNEXPECTED OBSERVATIONS are unique observations not collected in rigorous way but still intriguing and could inspire new investigations
- CONCLUSION brief statement as summary.
The organization of your discussion section is not fixed but rather it is driven by the reliability of the data you collect. The discussion should complement the logic set up with your biological rationale in the Introduction.
The following is not an appropriate discussion section: “Our data supported the hypothesis. The results were what we expected (see Results section).” Instead, state specifically what you observed in your data, and the conclusions you feel confident you can make based on the evidence you gathered. The Discussion should formulate and support a logical argument, leading the reader through the specific conclusions drawn from the data to their more general implications beyond the experiment.
Elements in the Discussion Section
Broad Study Question
What is the broad question that your research is trying to address? State your question clearly in the opening paragraph.
Support or Reject Hypothesis:
- If you have conducted a manipulative study, restate your hypothesis and whether you support or reject your hypothesis referencing appropriate data. (Note that finding no difference between two treatments is a result).
- Critically evaluate your biological rationale, experimental design, data collection, and explicit/implicit assumptions throughout. After this evaluation, you should be able to support or reject your hypothesis….OR you may feel that you did not fully test your hypothesis after all. A key step here is to look at your controls and variation in your measurements. How much variation surrounds your controls? How reliable and accurate are your measurements?
- Special Note about Inconclusive results: You may find that you have very LOW RELIABILITY of results because of malfunction, error or confounding variables. In this case, you may still feel your hypothesis is true, but you were not able to test it as expected. Instead, report your results as inconclusive, describing why you could not test your hypothesis and how you would revise your investigation in future studies.
- IMPORTANT NOTE: finding no difference between treatments is NOT an inconclusive result–No difference is a very valid result that contributes to a conclusion for either supporting or rejecting a hypothesis!
 Philosophical Note: DO NOT USE THE WORD PROVE. You cannot “prove” your hypothesis correct or incorrect. Science cannot prove anything, it can only provide evidence to support or reject your hypothesis. Without getting too philosophical, the role of science is not to find proof but rather to move closer to truth by eliminating hypotheses that are not true. Therefore, you will not be ‘proving your hypothesis’, but rather supporting or rejecting your hypothesis given the construct of your experiment and the data you have gathered. If you have carried out a systematic observation and may have not posed a formal hypothesis but you can provide answers to the general questions you posed about the system, or describe the system moreprecisely based on the data you collected.
Philosophical Note: DO NOT USE THE WORD PROVE. You cannot “prove” your hypothesis correct or incorrect. Science cannot prove anything, it can only provide evidence to support or reject your hypothesis. Without getting too philosophical, the role of science is not to find proof but rather to move closer to truth by eliminating hypotheses that are not true. Therefore, you will not be ‘proving your hypothesis’, but rather supporting or rejecting your hypothesis given the construct of your experiment and the data you have gathered. If you have carried out a systematic observation and may have not posed a formal hypothesis but you can provide answers to the general questions you posed about the system, or describe the system moreprecisely based on the data you collected.
Interpreting Data: If you feel that your protocol allowed you to test your hypothesis,
- Interpret each piece of data presented in the results independently and evaluate the reliability of the data.
- Discuss how these data are similar (confirm) or contrast with what is reported in literature you presented in the introduction OR new literature you discovered after you completed your experiment. Explain the trends you feel are important to support your conclusion(s) and evaluate how this supports or contradicts the biological assumptions you outlined in your biological rationale. Be prepared to detach yourself from your original biological rationale in explaining or being critical of your results.
- Combine and integrate the multiple types of reliable data you collected and discuss how together they inform the broad question (only combine data you are confident in).
Generating New Knowledge
Describe how your experiment contributes to the knowledge gap you identified in your introduction. Cite similar, contrary and/or supportive literature.
- If your data supported your hypothesis: guide your readers through the steps in your reasoning referring back to your biological rationale to provide context. Present the arguments that explain how your experimental approach and the pieces of evidence (data) convince you of your conclusion. Explain how do your findings add to those that others have observed. Compare your findings with information from the literature (this often requires a post-experimental literature search), citing appropriate references that support for your results. These references include many that you cited in your Introduction section; briefly summarize them but avoid redundancy.
- If your results are contrary to your hypothesis, you need to speculate the reasons for this difference, continue your literature search to explain your alternative results. Are your results consistent or inconsistent with others findings—why or why not? Distance yourself from the project while writing and be reasonably critical of your data. What evidence do you have that your biological rationale is acting? Is the mechanism you propose in effect? Evaluate the key biological assumptions in your biological rationale which were not correct.
- Implications of your findings- How does your experiment add to the current body of knowledge? Speculate on the implications of your findings. It is essential that you refer back to your biological rationale. Implications are specific, reasonable extensions of your results or the meaning of your results for the larger picture. Be careful, however, with your choice of words: state implications as logical possibilities rather than as fact. Your results may lead to new insights about relationships in nature. An unexpected result (if it holds up on repeating the experiment) may yield insight to guide a more effective experimental approach.
Evaluate Confidence in Experimental Design and Data Reliability/Quality
- Evaluate the strengths and weakness of your experiment and your confidence (or lack of) in your experimental design. Explain how these factors allow you to gauge the strength of your conclusion(s). Always address whether your protocol allowed you to truly test your hypothesis (see special note about inconclusive results in ‘support or reject hypothesis’ section above). In some cases you may discover unexpected inaccuracies in your data or that the methods you used were not appropriate or precise enough to address your question or test your hypothesis. Address the errors, unresolved issues and speculate how the experimental approach might be improved. Inconclusive results may show that you weren’t asking a relevant question in the first place or that the experiment was not able to test the question you posed. This, in turn, can generate specific new questions and experimental approaches. Avoid making a laundry list of mistakes you made in carrying out your experiment. Only mention errors if they help explain unexpected data values and/or lead you to conclude that your methods did not allow you to test your hypothesis.
- Evaluate reliability of data – Once you have established that your experimental design was appropriate to address your original question, you must also evaluate how well you carried out your intended design and what that means to your data reliability (e.g. evaluating whether the variation you see between samples is natural variation or experimenter’s error). How good are your data? Consider the variability in your data (variance, standard deviation, standard error). Did you have enough replicates? Did you have a large degree of experimental error? What are the implications of variability? Do not over-interpret your data. Recognize the magnitude of the variation within your data and the level of departure you would need to conclude true differences. In most cases you are trying to attach meaning to a group of numbers generated by some procedure. Help your readers make sense of these numbers by explaining how the patterns and relationships you observed reflect the biological concepts or issues you set out to explore. How do your data fit with your biological rationale?
New Questions and Future Studies: Science is built on an iterative cycle of questions, experiments, results and conclusions. Often it is appropriate to suggest the next step in the investigation. Be sure to include the reasoning that leads to your insights. Your experiment will likely provide many opportunities to ask new questions and suggest future studies.
Final Conclusion: End your paper strongly with a clear, brief conclusion that relates directly to the question, hypothesis, or knowledge gap you stated in the Introduction.
If you get stuck: The hard work of making meaning of data will be easier if you have a clear idea of what it was that you set out to do in the first place. Re-read your question and biological rationale. Do your results allow you to answer the question you posed in light of your biological rationale? A second reading of your BR after examining your data will often solve much of the confusion you may be experiencing. Be sure to discuss your results thoroughly with your research team. They may have some insight, intriguing literature for comparison, or thoughts about the data that could benefit your interpretation.
Other things you can do:
- Take a look at the example of good discussions on the next pages.
- Make a conceptual diagram for yourself or with your team. This is especially useful for seeing new connections, structuring ideas, and finding interactions at multiple levels.
- Explain the experiment and its significance to a friend who knows nothing about it. If you understand the full content, context, results and relevance of your experiment, you should be able to explain what was done and what it means. This should help provide some organization to your paper.
How will discussions be evaluated? To see our expectations for your Discussion, see the Biocore Research Paper Rubric in this Writing Manual.
Example of Good Discussion
Adapted from a paper by Jeremiah Wilke, Biocore 382, Fall 2003 Practice Paper entitled “Queen Anne’s Lace (Daucus carota) Species Frequency Suggests Rototilling as Most Effective method for Control of Invasive Weeds in Prairie Restoration Projects
The results suggest that rototilling is the most effective method as mulching and mowing yielded frequency values approximately 5 fold greater. The greater effectiveness of rototilling over the other methods coincides with previous knowledge of Queen Anne’s lace as it is known to favor habitats in no-till fields (Rose and Sheaffer, 2003) and re-sprout stems even after being cut (Biocore 382, class 2001, unpublished data) . (setting up logical argument: referring back to biological rationale and comparing findings with the literature). The frequency means suggest mowing to be slightly more effective than mulching; however, the distribution of the frequencies indicates little difference as the methods share common values. (Data interpretation- part of logical argument; Add re-statement of hypothesis and clearly state whether it was supported or rejected based on data interpretation)
Through rototilling seems to be the most efficacious for Queen Anne’s lace, several factors prevent us from making a definitive conclusion, most notably a small sample size. (Evaluating the validity and reliability of data) Frequency calculations can suggest patterns in the treatment, but they give no sense of the species density (number of a give species per quadrat). Examinations of the species frequency of Queen Anne’s lace in a control would also allow us to be more conclusive by gaining a sense of the improvement the methods made over untreated plots. (evaluating experimental design) Beyond our inability to decisively say which treatment is the most effective for Queen Anne’s lace, further work by the University of Wisconsin-Madison Biocore class of 2001 suggests we cannot generalize to other non-native species (Batzli, 2003). In their research, none of the methods demonstrated an appreciably greater capacity for weed control when tested on a variety of species. (discussion of other data makes our interpretation and argument more convincing) Species density calculations, measurements against a control, and the effectiveness of treatments on the other invasive plants therefore all necessitate future research. Mixing treatments has also been proposed (Batzli, 2003), while engineering novel methods deserves further study. (next steps)
(Final conclusion and brief discussion of implications of this research would help here)
Example of a Good Discussion that enumerates assumptions and how violating assumptions changes conclusions
Adapted from a poster by Beth Gausden, Katie Gielissen, Emily Gurnee, Jordan Mollet, and Carley Zeal, Biocore 384, Spring 2006
Addition of colchicines to MATa S. cerevisiae in vivo does not inhibit budding in the absence of α-factor but reduces shmooing and β-gal activity in response to α-factor
The results in Fig. 2 do not support our hypothesis (rejection of original hypothesis) that yeast exposed to colchicine in the absence of α-factor show a drastic decline in the incidence of budding as compared to controls. Our original hypothesis was based on the assumption that inhibition of mitotic division would prevent budding. (clear statement of key assumption in biological rationale) Although nuclear division is mediated by microtubules, pinching action and subsequent cytokinesis (budding) is controlled by actin filaments1. The tubulin-colchicine complex inhibits karygomy; however, bud formation can occur independently of nuclear division.1 Budding was still observed microscopically after three hours of incubation with colchicine (Fig. 2)- approximately two generations. These results indicate that bud formation was not inhibited by colchicine; (summary of how results do not support biological assumption) however, later generations incubated in colchicine may show complete cessation of budding as a result of aneuploidy, an irregular number of chromosomes.1 This occurs when a yeast cell undergoes successful cytokinesis but unsuccessful karyogamy; if this process is continuous or prolonged, cells will be unable to bud.
The results in Fig. 1 and Fig. 2 do not support our hypothesis that colchicine does not affect shmooing or the transcription of mating genes. We expected no change in the incidence of mating gene transcription as reported by the β-gal assay and percent of shmooing yeast in the yeast treated with colchicine compared to untreated yeast. The β-gal assay, Fig. 1, indicates a large decrease occurred in the transcription of mating genes in the presence of colchicine. Similarly, we observed a lower percentage of shmooing cells in the presence of colchicine. If nuclear division were inhibited by colchicine, then the portion of cells experiencing aneuploidy would be unable to respond to α-factor by shmooing or transcribing mating genes.
Conclusion
Our results suggest that colchicine does not inhibit bud formation (in the absence of α-factor) after 3 hours. We also observed decreased shmooing as well as β-galactosidase activity in yeast cells treated with colchicine and α-factor. The consistency of our results provides reasonable confidence in the methods. In future studies, longer incubation times, differing concentrations of colchicine, and chromosome and microtubule staining could be used to investigate the mechanism more thoroughly.
Example of Good Discussion
Adapted from a paper by Beth Theusch, Biocore 384, Spring 2003 Inorganic Phosphate Competitively Inhibits Alkaline Phosphatase-Catalyzed Hydrolysis of p-Nitrophenylphosphate
We hypothesized that inorganic phosphate (Pi) would act as a competitive inhibitor of the alkaline phosphatase-catalyzed pNPP hydrolysis reaction. Our data support this hypothesis. (re-statement of hypothesis and whether it was supported or rejected) As expected, we found that addition of inorganic phosphate increased the Km of the alkaline phosphatase-catalyzed pNPP hydrolysis reaction while the Vmax remained relatively unchanged. (setting up logical argument) After the addition of a concentration of Pi inhibitor approximately equal to the uninhibited Km substrate concentration, the apparent Km became 6-7 times as large (from 0.038 mM to 0.253 mM) as the uninhibited Km. Therefore, pNPP substrate molecules had to be almost 7 times as numerous as inhibitor molecules to access alkaline phosphatase’s active site and produce product equivalent to an initial uninhibited reaction velocity of 1/2 Vmax. These data indicate that Pi is quite an effective competitive inhibitor. One reason for its effectiveness as an inhibitor could be that the molecular weight (MW) of inorganic phosphate is about 96 g/mol, while the MW of pNPP, with its bulky nitrophenyl group, is almost 217 g/mol. Temperature is a measure of average molecular kinetic energy and is proportional to mv2. This means that lighter molecules have to move faster than heavy ones at 37oC in order to have the same kinetic energy as the large molecules. Molecules that move faster have more collisions, so it is likely that each Pi molecule had a greater chance of colliding with the alkaline phosphatase (AP) active site than did each pNPP substrate molecule during our experiment. (constructing new knowledge: references would help a lot here to show that the differences in molecular weight mentioned could significantly change kinetic energy) In addition, AP may have had a greater affinity for Pi than it did for the pNPP substrate, since alkaline phosphatases have a high affinity for inorganic phosphate (McComb et al., 1979). The bulky phenyl group on pNPP may have sterically hindered the hydrolysis reaction more than the hydrogen on Pi, depending on the specific geometry of the active site. As we mentioned previously, AP generally hydrolyzes Pi at a slower rate than it hydrolyzes phosphomonoesters (Schwartz, 1963), and so it may be that Pi occupies the AP active site longer per hydrolysis and thus excludes available pNPP from subsequently binding. (constructing new knowledge: referring back to biological rationale and comparing findings with the literature)
At first glance, it might appear that some of the increase in apparent Km could be attributed to a slight change in pH, since the Km value is pH dependent. Dibasic Pi can act as a base by adding a proton and becoming h1PO4- and as an acid by losing a proton and forming PO43-, but phosphate is predominantly the dianion at a pH of 8.6. Since the pH of the 0.05 mM Na2HPO4 salt solution was 7.7, which is close to the targeted value of 8.6, it is a reasonable to assume that the buffer counteracted any fluctuations in pH and essentially kept the pH constant. (evaluating experimental design)
Although the Vmax did not change dramatically between uninhibited and inhibited reactions, there was some difference between the uninhibited value of 0.056 umol/min and the inhibited value of 0.070 umol/min. Since Vmax did not decrease, it was clear that Pi did not act as a noncompetitive inhibitor. Since Vmax increases in the presence of an activator, it is possible that slight changes in ionic strength resulting from the addition of the salt could have activated AP somewhat. However, previous studies at a pH of 10 have shown that the activities of mammalian alkaline phosphatases are either unaffected or diminished by an increase in ionic strength. Specifically, calf intestinal AP experienced no change in activity following the addition of 1M NaCl, a much higher concentration than the Na+ that we introduced in our experiment. In other systems, NaCl addition at a pH of 9.0, close to the 8.6 we used in our experiment, had little effect on maximum velocity and actually inhibited it at low substrate concentrations (McComb et al., 1979). Since other variables in the experiment were held constant, the differences in Vmax values could simply be due to experimental error. (evaluating data reliability & experimental design)
The Ki value of 8.78 uM obtained from this study was comparable to but slightly greater than literature values for the Ki of E. coli AP. The values of 1 uM (O’Brien and Herschlag, 2001) and 0.6 uM (McComb et al., 1979) for Pi inhibition of E. coli AP were both obtained at a pH of 8.0 and temperature of 25oC, while we used a pH of 8.6, a temperature of 37oC, and bovine intestinal AP in our study. Just like Km values, Ki values are pH dependent. It is generally recognized that competitive inhibitors of AP are more effective at lower pHs (McComb et al., 1979). The pH difference alone could probably explain why our Ki was slightly larger and our inhibitor was slightly less effective than in the E. coli studies. In addition, bovine intestinal AP has a structure that is somewhat different from E. coli AP, so it is reasonable that the kinetics of the two enzymes could differ slightly. Some studies in rats have shown that only 1/10 as much Pi is needed to inhibit intestinal AP as compared to the amount that is needed to inhibit AP in other rat tissues (McComb et al., 1979). (evaluating data reliability & experimental design) Perhaps there are lower Pi concentrations in intestinal cells as compared to cells in other tissues. It would be interesting to see if this is true for bovine and other mammalian AP as well. (New questions/Future Studies)
The inhibition of AP by Pi, the product of AP catalyzed hydrolysis reactions, is a substrate-level regulation mechanism (Becker, Kleinsmith, and Hardin, 2003). This allows the AP enzyme to be responsive to product concentrations, so it is not always functioning at its maximum rate. It is not in the best interest of the cell to convert all phosphomonoesters into Pi and an alcohol at once, and the competitive inhibition by Pi helps to prevent this. This is precisely why initial reaction velocities are used when studying enzyme kinetics; if products are allowed to accumulate, they are likely to have an inhibitory effect on the enzyme. (implications of results, referring back to biological rationale)
Overall, the results of this study indicate that Pi is indeed a competitive inhibitor of bovine intestinal AP, as we had hypothesized. Specifically, we found that the Km value increased from 0.038 mM to 0.253 mM while Vmax remained relatively constant. We also found that our Ki value of 8.78 uM was reasonably similar to that reported previously for this particular enzyme and inhibitor. (final conclusion)
Parenthetical Citations Within Text
- Cite all information that you use from published or unpublished sources in the body of your paper and provide full citations in the Literature Cited section at end of the paper.
- Parenthetical author-date format within a sentence or at the end of a block of text. Provide the last name of the author(s) and the date the work was published, both enclosed by parentheses. Example: Global warming is a looming threat to biodiversity (Peters and Lovejoy 1992).
- More than one source, list them in chronological order: e.g. (Jones 1992; Smith and Jacobs 1993; Torrez 1995). If a work has more than two authors, you may list the first followed by et al. (latin for “and others”) and the date: (Jones et al. 1995). However, the names of all of the authors must be included in the list of citations at the end of the paper.
- Unpublished information: If you cannot find a published citation you can site personal communication in the body of your text – NOT in the literature cited. The format for unpublished information or data communication to you by a colleague is the source followed by “personal communication” or “unpublished data”: e.g. (Maria Rodriguez, personal communication 2002; Biocore 382 class, unpublished data). ***Use these sparingly as sources usually are not formal and cannot be verified easily. DO NOT base the major foundation of your study on personal communication unless the information gained is unique and not found elsewhere.
Literature Cited
List all works cited in the text – and no others – alphabetically in the References section at the end of your paper. The specific format used for references varies depending on each journal’s conventions, web-site format and the type of source to which you are referring. We would like you to use the format demonstrated below which follows the Name-Year system. Each reference should include the names of all the authors, the date the article or book was published and/or the date the website was accessed and its title. Regardless of the exact format used, make sure that you are consistent!
Here are some examples to follow:
Journal
Format as follows:
Author(s). year of publication. Title of the article (with only the first word capitalized). title of journal plus volume (issue): Inclusive page numbers.
One author example
Vitousek, P.M. 1994. Beyond global warming: ecology and global change. Ecology 75: 1861-1876.
Multiple author example
Post, W.M., Emanuel, W.R., Zinke, P.J., and Stangenberger, A.G. 1982. Soil carbon pools and world life zones. Nature 298: 156-159.
Internet Sources
A full discussion of number and types of internet resources is beyond the scope of this manual.
However, the following is a general guide for most articles that are published on the internet. As with all resources, especially those found on the internet, you must be wary of the source and its validity. If it doesn’t have an author or publication/ posting date BEWARE!
Format as follows:
Author(s). Year of publication. Title of the work. Title of the complete work or website or on-line journal plus volume (issue) if available/ applicable. Website URL or address (except for online journal or personal email). Date you accessed the web page.
Email:
Carbon, J.J. Physiology data. Personal email (7 July 2010).
Listserv or RSS feed newslist:
Blystone, R.V. 1994. Setting up a digital classroom and other stuff. biolab@hubcap.clemson.edu (accessed May 10, 1996).
World Wide Web: Basic form is: Author. Date. Title. URL (Access date)
Waterman, M., Stanley, E., Soderberg, P., and Jungck, J.R. 1999 Kingdoms entangled: molecules, malaria, and maise. BioQUEST Curriculum Consortium. http://bioquest.org/case.html (accessed April 12, 2012)
Macreal, H. 2001. Large Fish, Small Pond. http://www.bigfish.org/articles (accessed April 20, 2001)
Splice, G. 2000. Mutations are the Ultimate form of Variation. University Press Weekly vol 22. Electric Library. http://www.elibrary.com/ (accessed October 17, 2011).
*Note: Do not write out a website address (URL) as a parenthetic citation within the text of your paper—instead include the author and year of publication (e.g. Macreal 2001), just as you do with all other publications. Whenever possible, list the author. If you can’t find an author, list the organization that provided the information. If you can’t find the name of the organization, question the quality of your source.
Biocore Lab Manual
You will be citing one of your Biocore lab manuals in many of your research papers. To do this, look at the lab manual chapter to find the author(s) you wish to cite and the example format below. NOTE: This is an example for the Biocore Prairie chapter of the Biocore 382 lab manual.
Book Citations
Format as follows:
First author’s last name, First initials, subsequent authors’ name separated by commas, year of publication, title of book (italicized, with only the first word capitalized), edition number (if it is not the first edition), the publisher, the city of publication, and the state (omit the state for well known cities like New York).
Whole Book
Kuhn, T.S. 1962. The structure of scientific revolutions. University of Chicago Press, Chicago.
Purves, W.K., Sadava, D., Orians, G.H., and Heller, H.C. 2001. Life, the science of biology, 6th ed. Sinauer, Sunderland, MA.
Chapter in a Book
Naes, A. 1986. Intrinsic value: will the defenders of nature please rise? In Soulé, M.E., editor. Conservation biology: the science of scarcity and diversity. Sinauer Associates, Sunderland, MA. pp. 504-515.