Lab 1: PROCEDURE Part 4
PART 4: DESIGNING MUTATIONS IN HCAII (1 hr 15mins)
Mutagenesis of HCAII active site (30 min)
Now it is time to model mutating the protein!
- Close PyMOL, reopen it, and load the first session you saved: 2VVA.pse
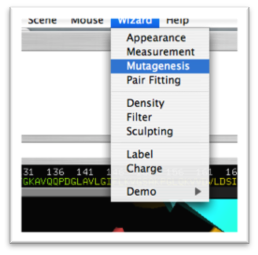
- To make a mutation, go to the menu: Wizard>Mutagenesis (Fig. 1.4)
- On the bottom of the side panel you will see the mutagenesis options. Click on No Mutation and select the amino acid type you would like to mutate a side chain to (Fig. 1.5).
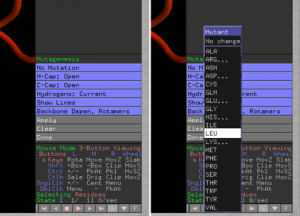
- Click on the side chain you would like to mutate, and the new amino acid type will appear in grey on top of the old one (Fig. 1.6). You can change the conformation of the new amino acid using the arrow buttons on the bottom-right corner.
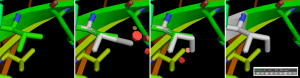
- You will see some red disks appearing. These are the potential clashes (atoms going into other atoms). A few small disks mean mild clashes, which are likely to be relieved by the protein (the backbone and the side chains will have some flexibility that is not present in your rigid model). Big disks (and many of them) means severe potential clashes.
NOTE: PyMOL has a problem mutating amino acids modeled in two conformations, such as His64. The mutation tool will not work for this residue. You can still propose a mutation here, but you won’t be able to visualize it.
Potential clashes are just potentials; predicted by PyMOL. They can help you to form hypothesis on how the mutations affect HCA II, but their actual effects have to be evaluated experimentally.
- Take a picture of your mutation. Save the mutation as a separate PyMOL session file (for example, mutation1.pse).
- Reload the 2VVA.pse PyMOL session and work on the next mutation.
You should select three point mutations in the active site and explain the rationale behind each mutation. You may, for example:
- Change the proton-shuttling His64 to a different amino acid. As noted above, you can’t visualize mutations to this residue using the mutagenesis tool, but you could still take a look at the residue and think about interesting mutations.
- Change the polarity of the cavity (hydrophobic-to-polar mutations or vice versa), while preserving the size
- Change a smaller residue to a larger one in a way that can be accommodated by the structure
- Purposely create clashing mutations that will likely compromise the structure
- Try to make a mutation that would increase HCAII catalytic activity
- Make mutations that will interfere with the position of the CO2, or otherwise change the shape of the active site
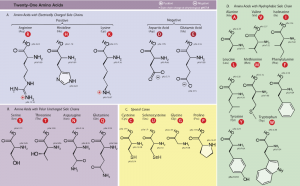
- Use figure 1.7 as a reference for choosing a mutation. Make sure to comment on how similar (or dissimilar) are the structures of the original residue and the proposed mutation (size/number of atoms, polarity, charge, etc.).
Effects of the mutations on DNSA binding (15 min)
- Open a new PyMOL session.
- Fetch PDB 1OKL, of HCAII complexed with dansylamide.
- Select the ligand by name: ‘sele resn MNS’ and save the selection.
- Consider the mutations you are proposing. Do you expect the binding of the dansylamide to be affected? Are the mutations compatible, or will they clash or generate a void? Be prepared to address these issues in your mini lab report.
Class presents the mutations and votes (30 min)
Find someone sitting near you and discuss the mutations you each made. Decide on your favorite one (from your total of six) to present to the class.
- Discuss your mutations with a partner. Between the two of you, choose one to present to the class. Once you’ve decided, write it up on the board. If someone else has already chosen your mutation, choose a different one.
- Create a Google slide in the class document to present your mutation and your rationale to the class. Be sure to include a clear image of your mutation and where it is relative to the active site. Address why you think this mutation is interesting and what effects you expect to see on the enzyme.
- A TA or volunteer will write the mutations on the board, along with a brief summary of the rationale.
- After every group has made a suggestion, the class will vote by a show of hands. Teaching staff will compile the mutations with the most votes. Ultimately, six of the mutants will be made, expressed and analyzed in the lab.
