53 An Introduction to Intermolecular Forces (M10Q1)
Introduction
In Modules 8 and 9, the attractive forces between atoms within an individual molecule (covalent bonds) has been examined. This section explores intermolecular forces, a collective term for attractive forces that occur between molecules, ions and non bonded atoms in bulk samples. The different types of intermolecular forces that exist are introduced, as well as how to determine what types of intermolecular force are important in a given substance or system.
Learning Objectives for Intermolecular Forces
- Observe that intermolecular forces exist.
| Intermolecular Forces | - Distinguish between intra- and intermolecular forces.
| Intramolecular Forces vs Intermolecular Forces | - Analyze a molecular structure to determine the appropriate intermolecular forces (eg. Ionic, Ion – Dipole, Dipole – Dipole, Hydrogen Bonding, Dipole − Induced Dipole, London Forces)
| Dispersion Forces | Dipole – Dipole Attractions | Hydrogen Bonding | Dipole – Induced Dipole Interactions | Ion – Dipole Interactions | Ionic Interactions | - Recognize the special role of H-bonds in water
| The Special Role of Hydrogen Bonds in Water |
| Key Concepts and Summary | Glossary | End of Section Exercises |
As was the case for gaseous substances, the kinetic molecular theory may be used to explain the behavior of solids and liquids. In the following description, the term particle will be used as a collective term to refer to an atom, molecule, or ion. Note that we will use the popular phrase “intermolecular attraction” to refer to attractive forces between the particles of a substance, regardless of whether these particles are molecules, atoms, or ions.
Consider these two aspects of the molecular-level environments in solid, liquid, and gaseous matter:
- Particles in a solid are tightly packed together and often arranged in a regular pattern; in a liquid, they are close together with no regular arrangement; in a gas, they are far apart with no regular arrangement.
- Particles in a solid vibrate about fixed positions and do not generally move in relation to one another; in a liquid, they move past each other but remain in essentially constant contact; in a gas, they move independently of one another except when they collide.
The differences in the properties of a solid, liquid, or gas reflect the strengths of the attractive forces between the atoms, molecules, or ions that make up each phase. The phase in which a substance exists depends on the relative extents of its intermolecular forces (IMFs) and the kinetic energies (KE) of its molecules. IMFs are the various forces of attraction that may exist between the atoms and molecules of a substance due to electrostatic phenomena, as will be detailed in this module. In this section, we will use the phase of a substance as an approximation of its IMFs, with solids having the strongest IMFs and gases having the weakest. These forces serve to hold particles close together, whereas the particles’ KE provides the energy required to overcome the attractive forces and thus increase the distance between particles. Figure 1 illustrates how changes in physical state may be induced by changing the temperature, hence the average KE, of a given substance.
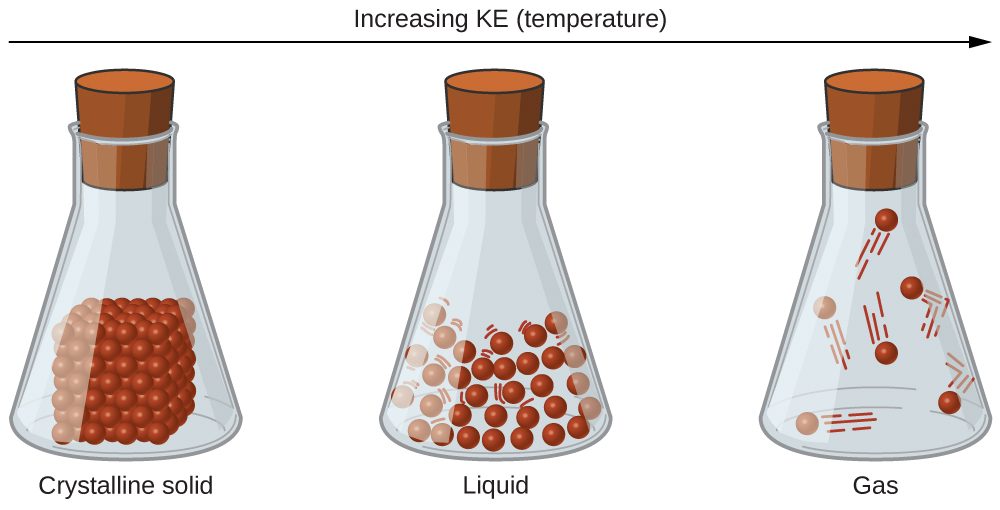
As an illustration of a process depicted in this figure, consider a sample of water vapor. When gaseous water is cooled sufficiently, by coming into contact with a cold surface for example, the attractions between H2O molecules will be capable of holding the molecules together when they come into contact with each other; the gas condenses, forming liquid H2O. For example, liquid water forms on the outside of a cold glass as the water vapor in the air is cooled by the cold glass, as seen in Figure 2.

Finally, if the temperature of a liquid becomes sufficiently low, or the pressure on the liquid becomes sufficiently high, the molecules of the liquid no longer have enough KE to overcome the IMF between them, and a solid forms.

Access this interactive simulation on states of matter, phase transitions, and intermolecular forces. This simulation is useful for visualizing concepts introduced throughout this module.
Intramolecular Forces vs. Intermolecular Forces
Intermolecular attractive forces are present in all substances as evidenced by the fact that everything will eventually solidify under appropriate conditions. They may be very weak, as is the case with the noble gases, but eventually even these atomic substances will liquify and then freeze if the temperature drops low enough. This is a result of the intermolecular forces as opposed to intramolecular forces. Intramolecular forces are those within a molecule that keep the atoms bonded together as an integral molecular unit. Intermolecular forces are the attractions between molecules or non bonded atoms, which determine many of the physical properties of a substance. Figure 3 illustrates the distinction between these two.
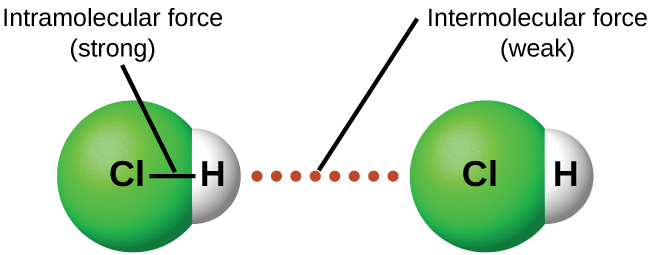
The strengths of IMFs vary widely, though usually the IMFs between small molecules are weak compared to the intramolecular forces that bond atoms together within a molecule. For example, to overcome the IMFs in one mole of liquid HCl(l) and convert it into gaseous HCl(g) (a physical change) requires only about 17 kilojoules. However, to break the covalent bonds between the hydrogen and chlorine atoms (a chemical change) in one mole of HCl requires about 25 times more energy—430 kilojoules. Attractive forces between neutral atoms and molecules are known collectively as van der Waals forces, although they are usually referred to more informally as intermolecular attraction. We will consider the various types of IMFs in the next sections of this module.
Dispersion Forces
The presence of this transient dipole can, in turn, distort the electrons of a neighboring atom or molecule in the sample, producing an induced dipole in the other molecule. Another name for this type of force is instantaneous dipole-induced dipole, though this term is less commonly used in our course. These two rapidly fluctuating, temporary dipoles thus result in a relatively weak electrostatic attraction between the species—a so-called dispersion force like that illustrated in Figure 5.
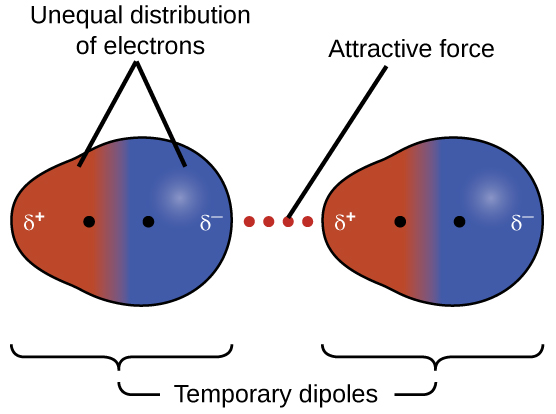
Dispersion forces that develop between atoms in different molecules can attract the two molecules to each other. The forces are relatively weak, however, and become significant only when the molecules are very close. Larger and heavier atoms and molecules exhibit stronger dispersion forces than do smaller and lighter atoms and molecules. F2 (38 g/mole) and Cl2 (71 g/mole) are gases at room temperature (reflecting weaker attractive forces); Br2 (160 g/mole) is a liquid, and I2 (254 g/mole) is a solid (reflecting stronger attractive forces). We will discuss this more in the next section on boiling points and melting points..
In a larger atom, the valence electrons are, on average, farther from the nuclei than in a smaller atom. Thus, the electrons are less tightly held and can more easily form the temporary dipoles that produce this attraction. The measure of how easy or difficult it is for another electrostatic charge (for example, a nearby ion or polar molecule) to distort a molecule’s charge distribution (its electron cloud) is known as polarizability. A molecule that has a charge cloud that is easily distorted is said to be very polarizable and a bulk sample will have relatively strong dispersion forces; one with a charge cloud that is difficult to distort is not very polarizable and a bulk sample will have relatively weak dispersion forces.
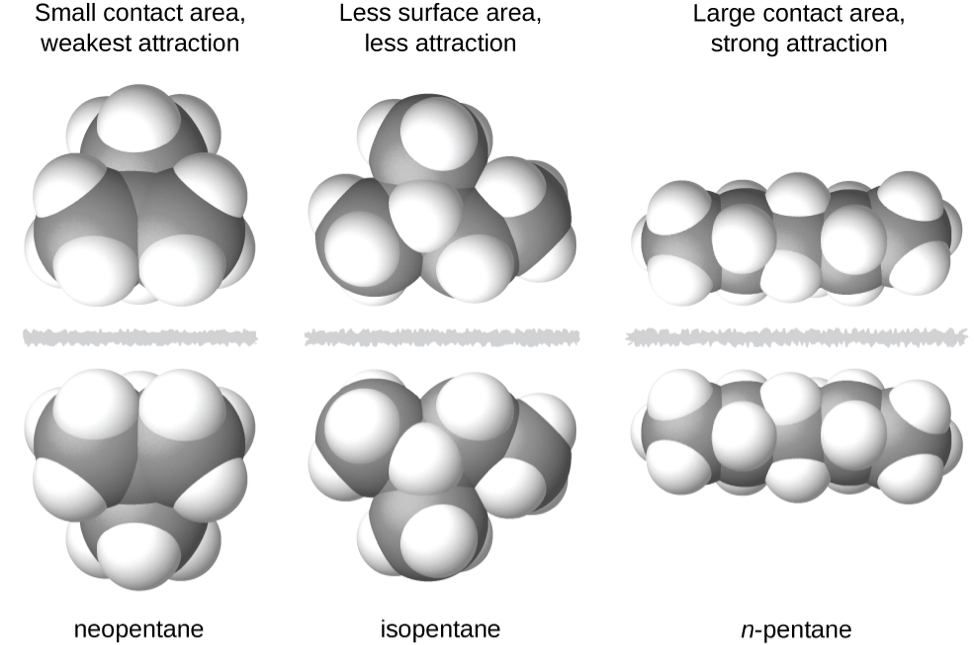
The shapes of molecules also affect the magnitudes of the dispersion forces between them. For example, consider the isomers n-pentane, isopentane, and neopentane (shown in Figure 6). Even though these compounds are composed of molecules with the same chemical formula, C5H12, the dispersion forces in the liquid phase are different, being greatest for n-pentane and least for neopentane. The elongated shape of n-pentane provides a greater surface area available for contact between molecules, resulting in correspondingly stronger dispersion forces. The more compact shape of isopentane offers a smaller surface area available for intermolecular contact and, therefore, weaker dispersion forces. Neopentane molecules are the most compact of the three, offering the least available surface area for intermolecular contact and, hence, the weakest dispersion forces. This behavior is analogous to the connections that may be formed between strips of VELCRO brand fasteners: the greater the area of the strip’s contact, the stronger the connection.
Chemistry in Real Life: Geckos and Intermolecular Forces
Geckos have an amazing ability to adhere to most surfaces. They can quickly run up smooth walls and across ceilings that have no toe-holds, and they do this without having suction cups or a sticky substance on their toes. And while a gecko can lift its feet easily as it walks along a surface, if you attempt to pick it up, it sticks to the surface. How are geckos (as well as spiders and some other insects) able to do this? Although this phenomenon has been investigated for hundreds of years, scientists only recently uncovered the details of the process that allows geckos’ feet to behave this way.
Geckos’ toes are covered with hundreds of thousands of tiny hairs known as setae, with each seta, in turn, branching into hundreds of tiny, flat, triangular tips called spatulae. The huge numbers of spatulae on its setae provide a gecko, shown in Figure 7, with a large total surface area for sticking to a surface. In 2000, Kellar Autumn, who leads a multi-institutional gecko research team, found that geckos adhered equally well to both polar silicon dioxide and nonpolar gallium arsenide. This proved that geckos stick to surfaces because of dispersion forces—weak intermolecular attractions arising from temporary, synchronized charge distributions between adjacent molecules. Although dispersion forces are very weak, the total attraction over millions of spatulae is large enough to support many times the gecko’s weight.
In 2014, two scientists developed a model to explain how geckos can rapidly transition from “sticky” to “non-sticky.” Alex Greaney and Congcong Hu at Oregon State University described how geckos can achieve this by changing the angle between their spatulae and the surface. Geckos’ feet, which are normally nonsticky, become sticky when a small shear force is applied. By curling and uncurling their toes, geckos can alternate between sticking and unsticking from a surface, and thus easily move across it. Further investigations may eventually lead to the development of better adhesives and other applications.
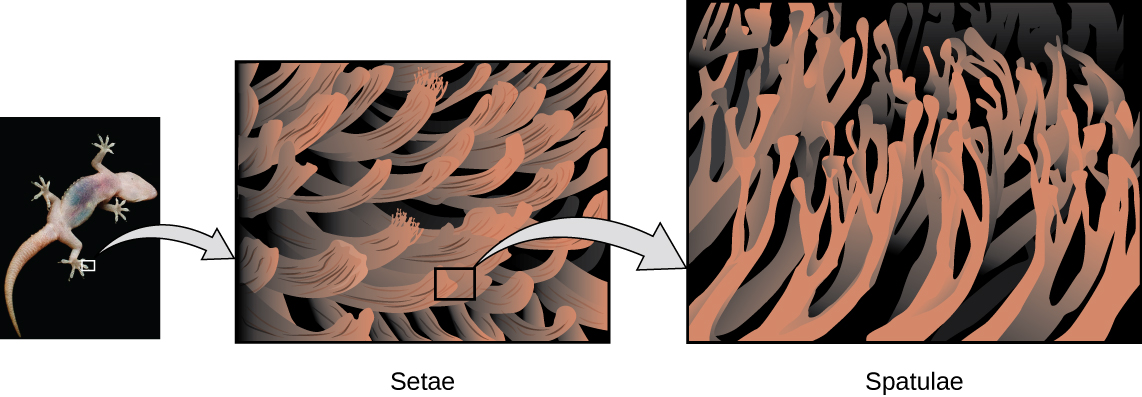

Read this article to learn more about Kellar Autumn’s research that determined that van der Waals forces are responsible for a gecko’s ability to cling and climb.
Dipole-Dipole Attractions
Recall from the module on Molecule Polarity that polar molecules have a partial positive charge on one side and a partial negative charge on the other side of the molecule—a separation of charge called a dipole. Consider a polar molecule such as hydrogen chloride, HCl. In the HCl molecule, the more electronegative Cl atom bears the partial negative charge, whereas the less electronegative H atom bears the partial positive charge. An attractive force between HCl molecules results from the attraction between the positive end of one HCl molecule and the negative end of another. This attractive force is called a dipole-dipole attraction—the electrostatic force between the partially positive end of one polar molecule and the partially negative end of another, as illustrated in Figure 8.
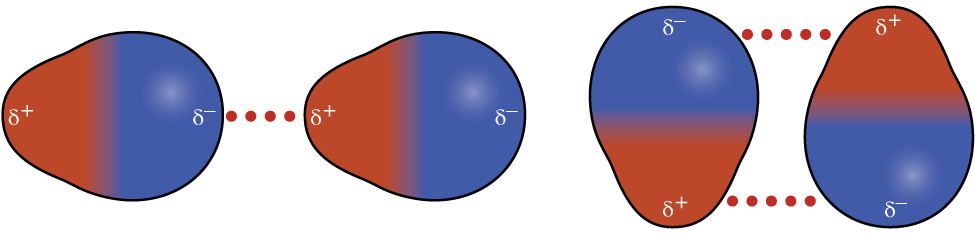
The effect of a dipole-dipole attraction is apparent when we compare the properties of HCl to nonpolar F2. Both HCl and F2 are diatomic molecules with approximately the same molar mass and therefore bulk samples of these substances would have comparable London forces. However, HCl(l) has a much higher boiling point (188 K) compared to F2(l) (85 K), an indication that HCl(l) has stronger IMFs as a result of dipole-dipole attractions in addition to the London forces.
Hydrogen Bonding
Nitrosyl fluoride (ONF, molecular mass 49 amu) is a gas at room temperature. Water (H2O, molecular mass 18 amu) is a liquid, even though it has a lower molecular mass. We clearly cannot attribute this difference between the two compounds to dispersion forces. Both molecules have about the same shape and ONF is the heavier and larger molecule. It is, therefore, expected to experience more significant dispersion forces. Additionally, we cannot attribute this difference to differences in the dipole moments of the molecules. Both molecules are polar and exhibit comparable dipole moments. The difference is due to a particularly strong dipole-dipole attraction that may occur when a molecule contains a hydrogen atom bonded to a fluorine, oxygen, or nitrogen atom (three of the most electronegative elements). The very large difference in electronegativity between the H atom (2.2) and the atom to which it is bonded (4.0 for an F atom, 3.5 for an O atom, or 3.1 for a N atom), combined with the very small size of a H atom and the relatively small sizes of F, O, or N atoms, leads to highly concentrated partial charges with these atoms. Molecules with F-H, O-H, or N-H groups are very strongly attracted to similar groups in nearby molecules, and a particularly strong type of dipole-dipole attraction called hydrogen bonding is formed. This hydrogen bond forms from the interaction between the lone pair of electrons on an N, O, or F atom (the donor) and the antibonding orbital in a highly polarized sigma bond (the acceptor). Examples of hydrogen bonds include HF⋯HF, H2O⋯HOH, and H3N⋯HNH2, in which the hydrogen bonds are denoted by dots.
Figure 9 illustrates hydrogen bonding between water molecules. Notice that the hydrogen bond forms between a hydrogen that is attached to an oxygen atom on one molecule and an oxygen atom on an adjacent molecule. This example highlights the two requirements for forming a hydrogen bond: (1) One molecule must have an H atom bonded to an N, O, or F atom; and (2) another molecule must have an N, O, or F atom with a lone pair of electrons.
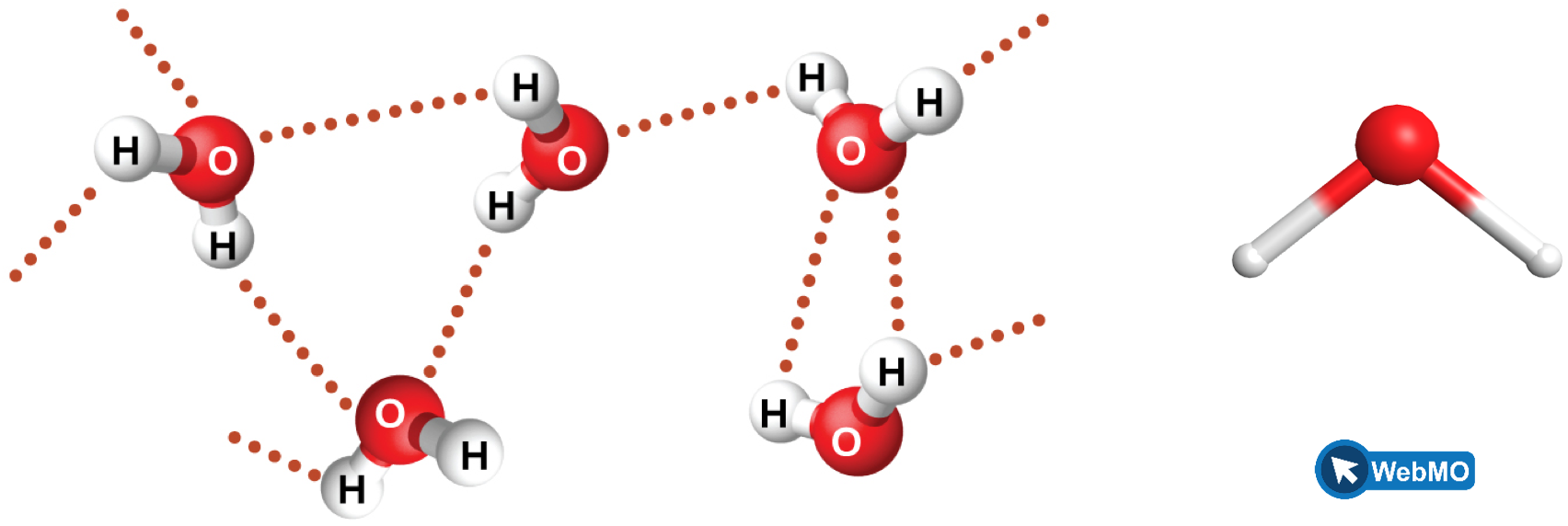
Despite use of the word “bond,” keep in mind that hydrogen bonds are much weaker than covalent bonds, only about 5 to 10% as strong, but are generally much stronger than other dipole-dipole attractions and dispersion forces. For example, hydrogen bonds have a pronounced effect on the properties of condensed phases (liquids and solids).
So far we have emphasized that IMFs are between separate molecules, but this is not always the case. In very large molecules such as proteins or DNA, these IMFs can occur within the same molecule and help stabilize their three-dimensional structure, which is essential for their mode of operation. The hydrogen bonding in DNA is described below.
Chemistry in Real Life: Hydrogen Bonding and DNA
Deoxyribonucleic acid (DNA) is found in every living organism and contains the genetic information that determines the organism’s characteristics, provides the blueprint for making the proteins necessary for life, and serves as a template to pass this information on to the organism’s offspring. A DNA molecule consists of two (anti-)parallel chains of repeating nucleotides, which form its well-known double helical structure, as shown in Figure 10.
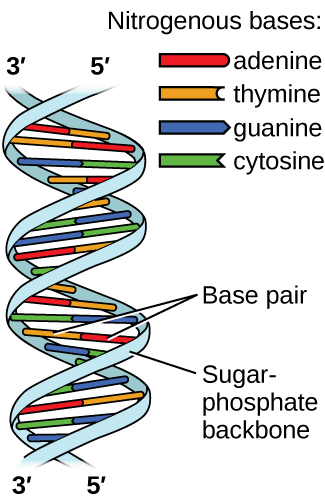
Each nucleotide contains a (deoxyribose) sugar bound to a phosphate group on one side, and one of four nitrogenous bases on the other. Two of the bases, cytosine (C) and thymine (T), are single-ringed structures known as pyrimidines. The other two, adenine (A) and guanine (G), are double-ringed structures called purines. These bases form complementary base pairs consisting of one purine and one pyrimidine, with adenine pairing with thymine, and cytosine with guanine. Each base pair is held together by hydrogen bonding. A and T share two hydrogen bonds, C and G share three, and both pairings have a similar shape and structure (Figure 11).
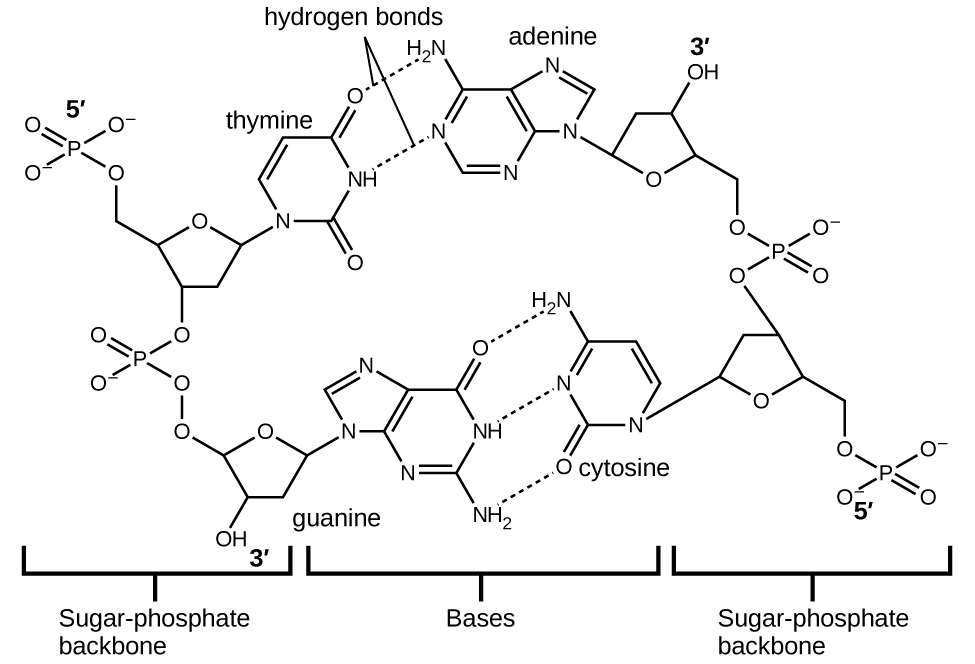
The cumulative effect of millions of hydrogen bonds effectively holds the two strands of DNA together. Importantly, the two strands of DNA can relatively easily “unzip” down the middle since hydrogen bonds are relatively weak compared to the covalent bonds that hold the atoms of the individual DNA molecules together. This allows both strands to function as a template for replication.
Dipole-Induced Dipole Interactions
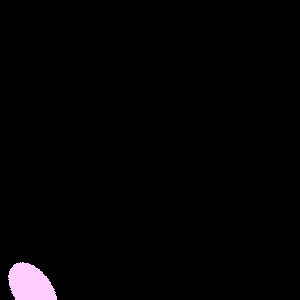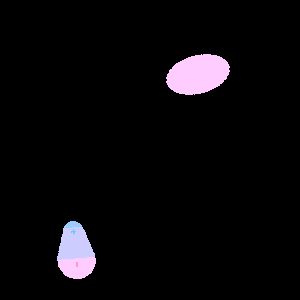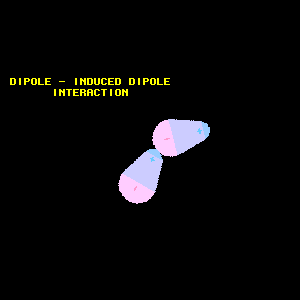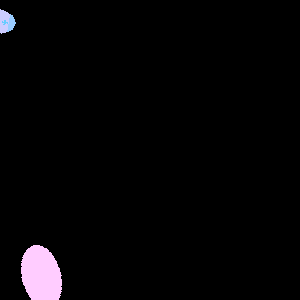
In the Dispersion Forces section, we learned that at any moment, an atom or nonpolar molecule can experience an instantaneous dipole if its electrons are asymmetrically distributed, and that this instantaneous dipole can induce a dipole in a neighboring atom or molecule. If a molecule with an established dipole were in proximity to a nonpolar molecule, however, it follows that the established dipole will induce a dipole in the nonpolar molecule. This type of interaction between a polar molecule and a nonpolar molecule is known as a dipole-induced dipole interaction (Figure 12).
Ion-Dipole Interactions
Ion-dipole interactions are the result of the Coulombic electrostatic interactions between an ion and the charged ends of a dipole. Since a dipole consists of an end that is slightly positively charged and another end that is slightly negatively charged, there will be Coulombic attraction and repulsion in this interaction. Consider a cation and a polar molecule. The positive-positive repulsion means the positive end of the dipole is going to be farther away from the cation than the negative end that is attracted (Figure 13).

Ionic Interactions
In Module 2, you learned about ionic compounds. Recall that when an element composed of atoms that readily lose electrons (a metal) reacts with an element composed of atoms that readily gain electrons (a nonmetal), a transfer of electrons usually occurs, producing a positively charged cation and a negatively charged anion. These ions then experience electrostatic attractions (ionic bonds) due to the ions of opposite charge present in the compound.
The Special Role of Hydrogen Bonding in Water
All substances become less dense when they are heated (as they expand slightly) and more dense when they are cooled. Above 4°C water is no exception, but below 4°C as it is cooled further, liquid water actually expands a little and the density decreases slightly. Upon actually freezing at 0 oC, there is a much larger expansion as the volume increases by about 9% with a corresponding drop in density as the liquid water becomes solid ice. In H2O(s) each water molecule makes four hydrogen bonds with its neighbors resulting in a three dimensional array with an open hexagonal structure, more open and therefore with a greater volume compared to the same amount of H2O(l). Between 4 oC and 0 oC, the water molecules are beginning to orient themselves and so the liquid expands as it gets close to actually freezing. (Figure 9). You have likely found that if you place a full water bottle in the freezer, the plastic bottle will begin to bulge as the solid water occupies more space than the liquid water.
The expansion of ice as it freezes means that ice floats on top of water. This characteristic is vital to the survival of organisms in cold climates like Wisconsin. During the winter, as Lake Mendota begins to freeze, the surface of the water freezes first; people often enjoy skating or skiing across frozen lakes when the ice is thick enough. The ecosystem is able to survive in the unfrozen water beneath the surface because the ice insulates it, providing an adequate living temperature. If water froze and sank to the bottom, lakes would freeze from the bottom up and become completely frozen during winter. The summer is not long enough in Wisconsin to melt all of the water in the frozen lakes, and fish and other organisms would be unable to survive.
Key Concepts and Summary
The physical properties of condensed matter (liquids and solids) can be explained in terms of the kinetic molecular theory. In a liquid, intermolecular attractive forces hold the molecules in contact, although they still have sufficient KE to move past each other.
Intermolecular attractive forces, collectively referred to as van der Waals forces, are responsible for the behavior of liquids and solids and are electrostatic in nature. Dipole-dipole attractions result from the electrostatic attraction of the partial negative end of one polar molecule for the partial positive end of another. The temporary dipole that results from the motion of the electrons in an atom can induce a dipole in an adjacent atom and give rise to the London dispersion force. London forces increase with increasing molecular size. Hydrogen bonds are a special type of dipole-dipole attraction that results when hydrogen is bonded to one of the three most electronegative elements: F, O, or N.
Glossary
- dipole-dipole attraction
- intermolecular attraction between two permanent dipoles
- dispersion force
- (also, London dispersion force or London force) attraction between two rapidly fluctuating, temporary dipoles; significant only when particles are very close together
- hydrogen bonding
- occurs when exceptionally strong dipoles attract; bonding that exists when hydrogen is bonded to one of the three most electronegative elements: F, O, or N
- induced dipole
- temporary dipole formed when the electrons of an atom or molecule are distorted by the instantaneous dipole of a neighboring atom or molecule
- instantaneous dipole
- temporary dipole that occurs for a brief moment in time when the electrons of an atom or molecule are distributed asymmetrically
- intermolecular force
- noncovalent attractive force between atoms, molecules, and/or ions
- polarizability
- measure of the ability of a charge to distort a molecule’s charge distribution (electron cloud)
- van der Waals force
- attractive or repulsive force between molecules, including dipole-dipole, dipole-induced dipole, and London dispersion forces; does not include forces due to covalent or ionic bonding, or the attraction between ions and molecules
Chemistry End of Section Exercises
- Open the PhET States of Matter Simulation to answer the following questions:
- Select the Solid, Liquid, Gas tab. Explore by selecting different substances, heating and cooling the systems, and changing the state. What similarities do you notice between the four substances for each phase (solid, liquid, gas)? What differences do you notice?
- For each substance, select each of the states and record the given temperatures. How do the given temperatures for each state correlate with the strengths of their intermolecular attractions? Explain.
- Select the Interaction Potential tab, and use the default neon atoms. Move the Ne atom on the right and observe how the potential energy changes. Select the Total Force button, and move the Ne atom as before. When is the total force on each atom attractive and large enough to matter? Then select the Component Forces button, and move the Ne atom. When do the attractive (van der Waals) and repulsive (electron overlap) forces balance? How does this relate to the potential energy versus the distance between atoms graph? Explain.
- Define the following and give an example of each:
- dispersion force
- dipole-dipole attraction
- hydrogen bond
- Explain why a hydrogen bond between two water molecules is weaker than a hydrogen bond between two hydrogen fluoride molecules.
- Identify the intermolecular forces present in the following solids:
- CH3CH2OH
- CH3CH2CH3
- CH3CH2Cl
Answers to Chemistry End of Section Exercises
- (a) Similarities: Solid and liquid are more dense compared to gas. The higher temperature the system has, the faster the molecules move. Differences: For water, the liquid state is denser than solid state; while other solids are denser than their liquid state. The various substances reach each state at different temperatures.
(b) Neon: -259 °C (solid), -246 °C (liquid), -217 °C (gas); Argon: -230 °C (solid), -187 °C (liquid), -84 °C (gas); Oxygen: -246 °C (solid), -216 °C (liquid), -160 °C (gas); Water: -127 °C (solid);, 13 °C (liquid), 156 °C (gas). Solids have the strongest IMFs and gases have the weakest. For each state, IMF decreases with increasing temperature.
(c) When the distance between two atoms is greater than ε , the total force on each atom attractive and large enough to matter. When the distance between two atoms is ε , the attractive (van der Waals) and repulsive (electron overlap) forces balance. At this point, the whole system has the lowest potential energy. - (a) Dispersion forces occur as an atom develops a temporary dipole moment when its electrons are distributed asymmetrically about the nucleus. This structure is more prevalent in large atoms such as argon or radon. A second atom can then be distorted by the appearance of the dipole in the first atom. The electrons of the second atom are attracted toward the positive end of the first atom, which sets up a dipole in the second atom. The net result is rapidly fluctuating, temporary dipoles that attract one another (example: Ar).
(b) A dipole-dipole attraction is a force that results from an electrostatic attraction of the positive end of one polar molecule for the negative end of another polar molecule (example: ICI molecules attract one another by dipole-dipole interaction).
(c) Hydrogen bonds form whenever a hydrogen atom is bonded to one of the more electronegative atoms, such as a fluorine, oxygen, or nitrogen atom. The electrostatic attraction between the partially positive hydrogen atom in one molecule and the partially negative atom in another molecule gives rise to a strong dipole-dipole interaction called a hydrogen bond (example: HF • • • HF). - The hydrogen bond between two hydrogen fluoride molecules is stronger than that between two water molecules because the electronegativity of F is greater than that of O. Consequently, the partial negative charge on F in HF is greater than that on O in H2O.
- (a) hydrogen bonding and dispersion forces; (b) dispersion forces; (c) dipole-dipole attraction and dispersion forces

