Unit Two
Day 16: DNA and Lipids
D16.1 DNA
Deoxyribonucleic acid (DNA) consists of two polymer chains that coil around each other, forming a double helix. The monomer units of DNA strands are called nucleotides. A nucleotide consists of a phosphate group and a nucleobase attached to a sugar (deoxyribose).
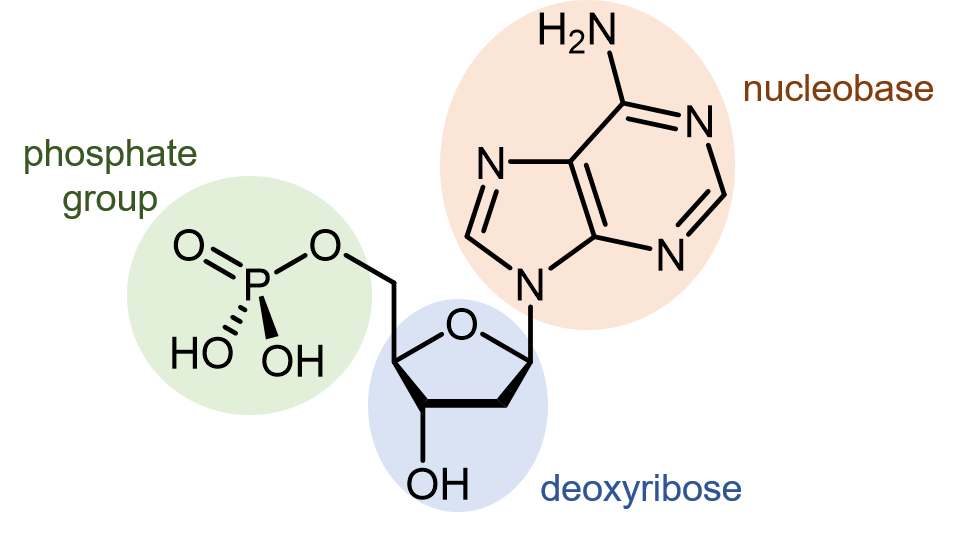
There are four different nucleobases found in DNA. They are called adenine (abbreviated A), thymine (T), guanine (G) and cytosine (C). The nucleotides containing those nucleobases are called deoxyadenosine monophosphate, deoxythymidine monophosphate, deoxyguanosine monophosphate, and deoxycytidine monophosphate (click on each name to see a rotatable molecular model).
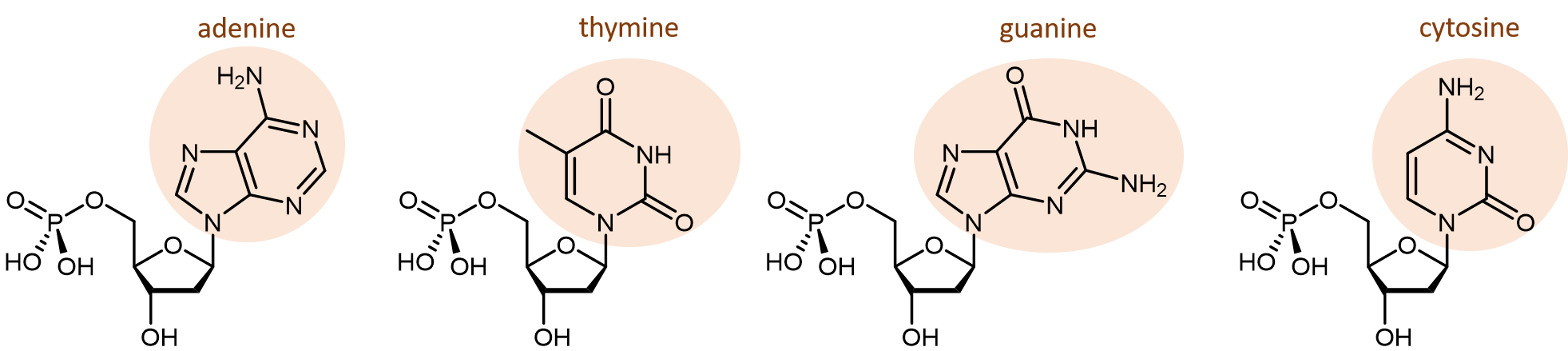
The polymerization reaction forming a DNA strand is a condensation reaction between the phosphate group and deoxyribose (specifically the -OH group), which results in the formation of phosphodiester bonds joining the nucleotides. In organisms containing DNA the pH is about 7, and at that pH the phosphate groups are ionized. Thus, DNA is a huge polyatomic anion with one negative charge for each monomer unit along the chain. The negative charge is balanced by positive charges on metal ions, protein molecules, and other cations surrounding the DNA.
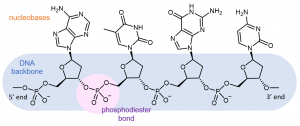
Each DNA polymer chain has a backbone of alternating sugar group and phosphodiester bond, with aromatic nitrogen-containing nucleobases forming side-chains bonded to the backbone.
A DNA strand has a free phosphate group at one end (called the 5′ end because the phosphate is attached to the carbon-5 position in the ribose) and a free hydroxyl (-OH) group at the other end (called the 3′ end because the hydroxyl is attached to the carbon-3 position in the ribose). By convention, the sequences are named from 5′ to 3′, and only the nucleobases are included in the name. For example, the molecule shown above is ATGC (not CGTA).
Exercise 1: Forces within DNA
Two single strands of DNA combine to make the double-helix molecular anion (typically called just the DNA molecule). The strands run in opposite directions: at one end of the double helix is the 5′ end of one strand and the 3′ end of the other strand. The two strands wrap around each other to form two intertwined helixes. The rotating figure shows how the two helixes are oriented. Note that one of the gaps between the entwined strands is wider than the other gap. The two separate strands are held in this orientation by hydrogen bonding between nucleobases in the center of the molecule.

D16.2 Base Pairing
One of the most remarkable things about DNA is that the quantity of adenine always equals the quantity of thymine and the quantity of guanine always equals the quantity of cytosine. From one organism to another, the quantity of adenine can vary, but the quantity of thymine always varies the same way. These equalities imply that the nucleobases occur in pairs: adenine paired with thymine and guanine paired with cytosine. These pairs of nucleobases are called complementary base pairs.
Out of six possible combinations, why are these two the complementary pairs? First, the nucleobases have different sizes: T and C have single rings and are smaller; A and G have double rings and are larger. If A and G paired, their larger size would force the double helix to bulge out. If T and C paired, their smaller size would cause pinching of the double helix. Only a pairing between a single-ringed and a double-ringed nucleobase has the proper consistent spacing.
This leaves four possible combinations: A-C, A-T, G-C and G-T. Of these, A-T and G-C pairs maximize the number of hydrogen bonds across the shared helical axis:
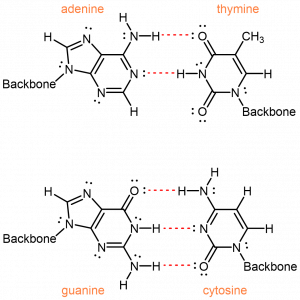
The A-C and G-T pairings, on the other hand, would have fewer hydrogen bonding interactions. To achieve any hydrogen-bonding, there would have to be awkward positioning of the nucleobases relative to each other.
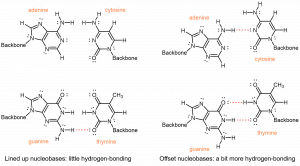
Therefore, intermolecular interactions, specifically the efficient hydrogen-bonding, between adenine and thymine, and between guanine and cytosine, result in the specificity observed for the complementary base pairs.
Because of the nature of complementary base pairing, if you know the sequence of one strand of DNA, you can predict the sequence of the strand that will pair with, or “complement” it.
During DNA replication, the strands of the double helix of DNA first separate. Then, enzymes catalyze the synthesis of new DNA in the 5′ to 3′ direction, using the two original strands as models, or templates, for the complementary strands. Two complete DNA molecules, each an exact copy of the original, are the result.
Because of the specificity of base pairing and a proofreading activity of the enzyme that synthesizes new DNA, less than one error in 100 million is incorporated into the new DNA strands. Thus genetic information is passed accurately on to the next generation.
Exercise 2: Forces within DNA Molecules
D16.3 Lipids
Fats and oils are part of a class of biomolecules called lipids, which are loosely defined as biomolecules that are insoluble in water but soluble in organic solvents like hexane or chloroform. Here, we discuss two important classes of lipids: glycerolipids and phospholipids. The molecular structures of lipids give rise to different intermolecular interactions that influence their physical properties and biological functions.
D16.4 Glycerolipids
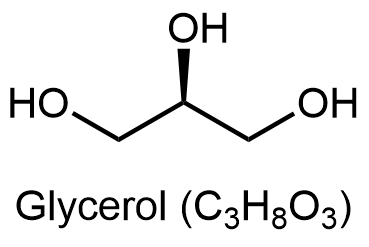
Glycerolipids are composed of glycerol and fatty acids. Glycerol has three carbon atoms, each of which has a hydroxyl (-OH) group bonded to it:
Fatty acids are long, unbranched hydrocarbon chains with a carboxylic acid group at one end.
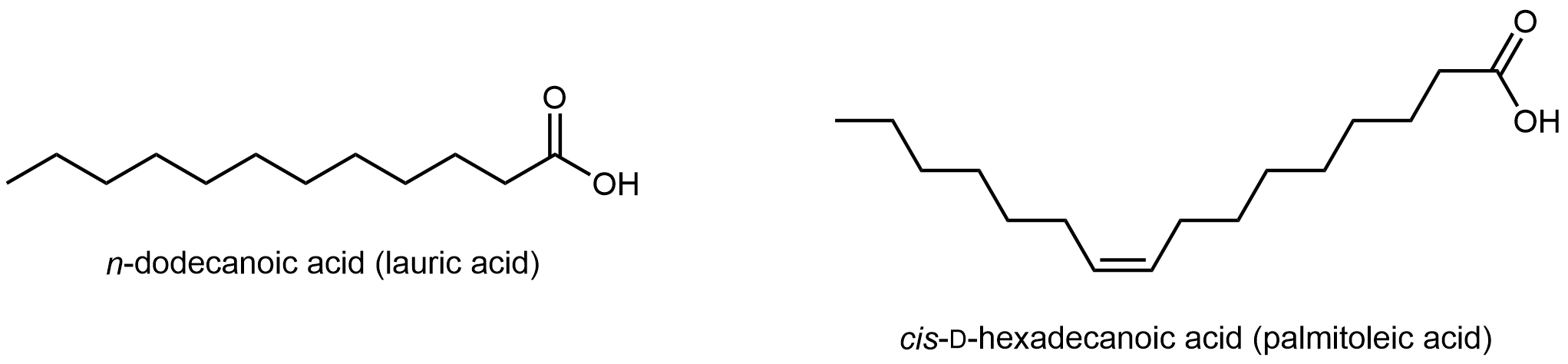
Fatty acids are systematically named based on the number of carbon atoms and C=C double bonds in the chain. But their common names, such as lauric acid, are often used to identify them.
Note that there are only C-C single bonds in lauric acid; similar to hydrocarbons, such a fatty acid is a saturated fatty acid. In contrast, palmitoleic acid has a C=C double bond, i.e. it has one degree of unsaturation. The π bond can undergo addition of H2 and form a saturated fatty acid. Lipids made from saturated and unsaturated fatty acids have different properties.
Glycerolipids are formed when three fatty acids are joined to glycerol. Catalyzed by enzymes, each hydroxyl group on glycerol can undergo condensation with the carboxylic acid group of a fatty acid, forming an ester linkage.
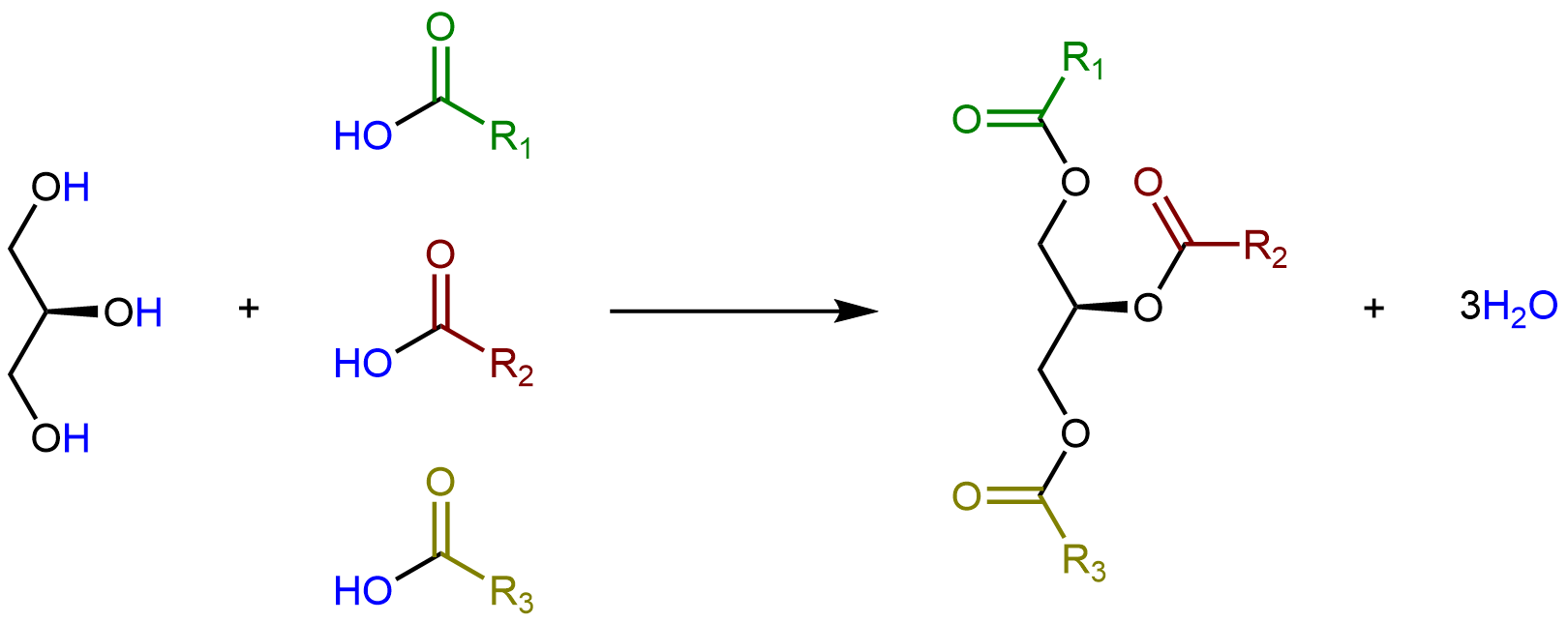
The lengths of the fatty acid chains (e.g. R1, R2, R3) and the number of C=C double bonds within them determine the melting point of a glycerolipid. Longer chains have stronger London dispersion forces than shorter chains, leading to a higher melting point. Saturated chains can pack against each other regularly, giving rise to stronger London dispersion forces between the chains and higher melting points. Unsaturated chains, especially with cis double bonds, which force the chain to bend, cannot stack together as well, so the London dispersion forces are weaker.
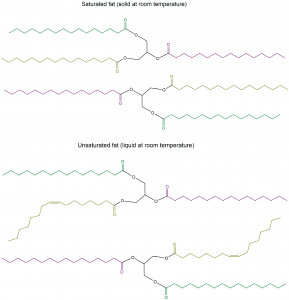
Glycerolipids with a higher melting point that are usually solids at room temperature are called fats or waxes. Glycerolipids with a lower melting point that are usually liquids at room temperature are called oils.
An important role of glycerolipids is in energy storage. The carbon atoms in fatty acids are mostly involved in C-H bonds, and therefore can be oxidized and in the process release energy. In living organisms, this is carried out in a controlled manner via enzymes. But we can also burn fats and waxes (for example, in a candle) to obtain energy in the form of light.
D16.5 Phospholipids
If one of the fatty acids in a glycerolipid is replaced by a hydrophilic phosphate group, the result is a phospholipid molecule.
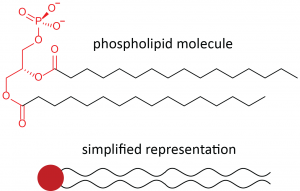
Below the line structure is a common, simplified way of representing a phospholipid molecule. The phosphate group, together with the polar ester groups, is often called the hydrophilic head of the molecule (red circle). The fatty-acid hydrocarbon chains are often called the hydrophobic tails of the molecule (black squiggly lines). Molecules with both hydrophobic and hydrophilic parts are called amphiphiles or amphiphilic molecules.
In an aqueous environment, such as a living cell, amphiphilic molecules spontaneously arrange into ordered structures. The most important of these is a lipid bilayer, two layers of phospholipid molecules with the hydrophobic tails intermingled and the hydrophilic heads contacting the water. As the diagram below shows, water is excluded from the center of the bilayer where the fatty acid tails are, while the polar head-groups have access to the water. Phospholipid bilayers form the basis for cell membranes and all the other organelle boundaries found in living cells. Membranes found in cells also contain many other components, such as free fatty acids, other lipids, and proteins.

Since the center of the bilayer is nonpolar, only nonpolar molecules, such as O2, CO and CO2, can cross the bilayer. Ions, such as K+, Na+, and Cl–, are completely blocked by bilayers. Because of the large number of atoms that would have to be rearranged to allow passage of large molecules, fatty acids, complex carbohydrates, proteins, and nucleic acids are also blocked by bilayers.
16.6 Proteins, Lipids, and Fatty Acids
Passage of large molecules or ions through lipid bilayers, can be controlled by protein molecules that are embedded in a bilayer and extend outside the bilayer on both sides. In these membrane-spanning proteins, side chains facing the lipid membranes are usually hydrophobic; side chains contacting the aqueous space on either side of the membrane can be polar.
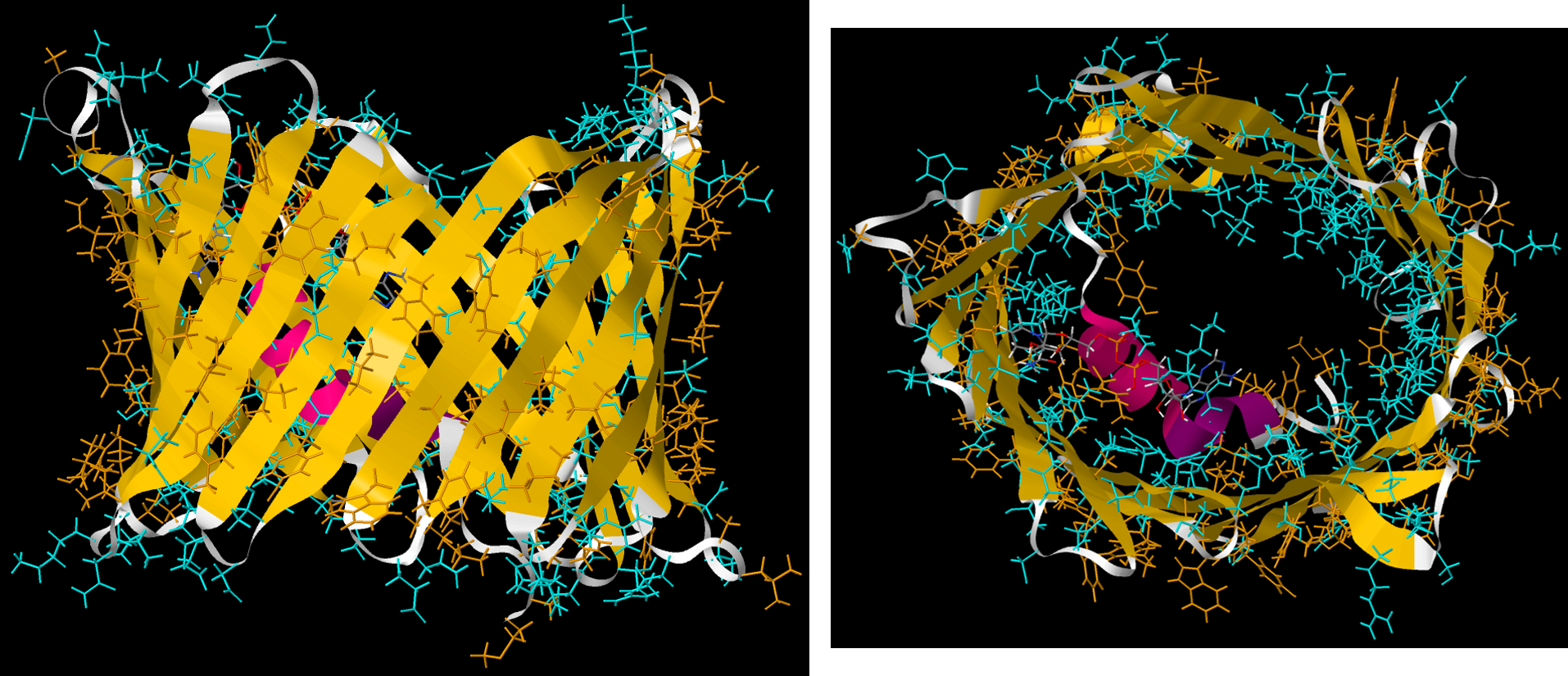
Many proteins are anchored to cell membranes by forming a covalent bond to a fatty acid. For example, the amine group (-NH2) on the N-terminus of a protein or in a lysine side chain can undergo condensation with the carboxylic acid group (-COOH) on a fatty acid found in a cell membrane to form an amide linkage. Such a covalent bond keeps the protein associated with the membrane.
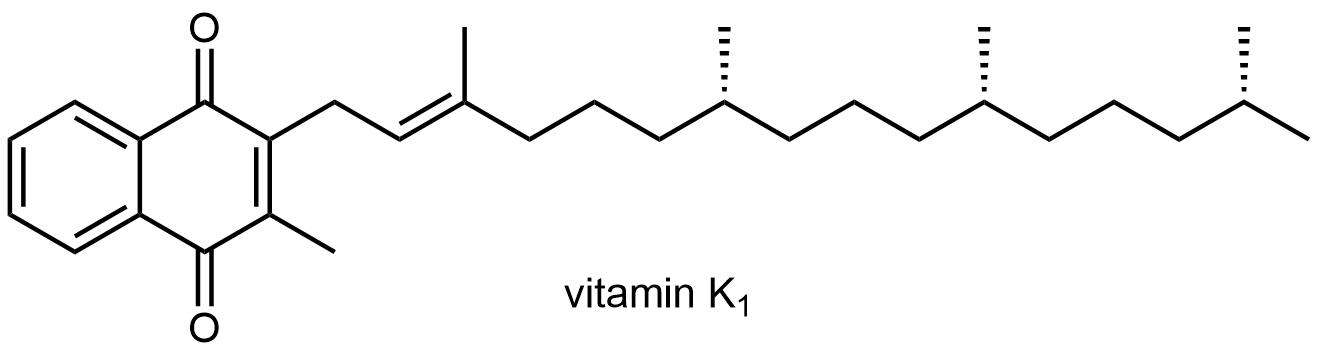
Several vitamins necessary for higher animal life are only soluble in lipids. These vitamins are composed almost entirely of carbon and hydrogen, meaning they are hydrophobic like the fatty acid tails of lipids. A diet completely lacking in fat will be deficient in these vitamins unless the vitamins are taken as pills.
For example, vitamin K1 is necessary for blood clotting:
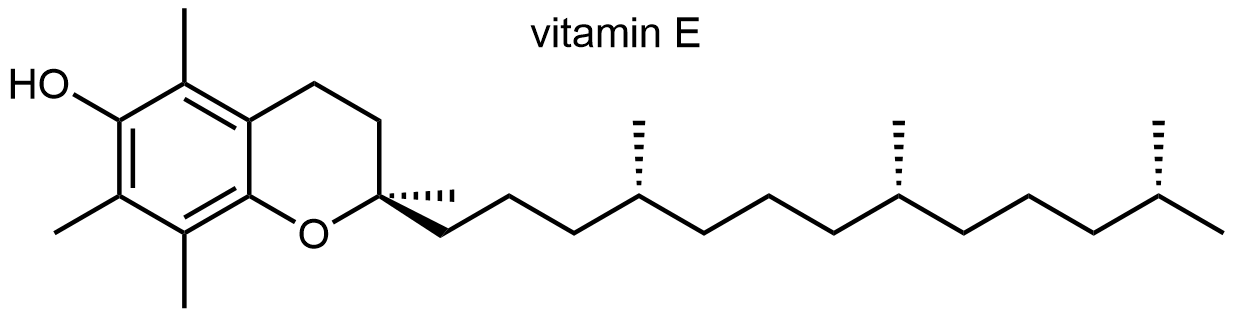
α-Tocopherol (Vitamin E) prevents the inappropriate oxidation of membrane lipids. Deficiency in Vitamin E also causes infertility in rats.
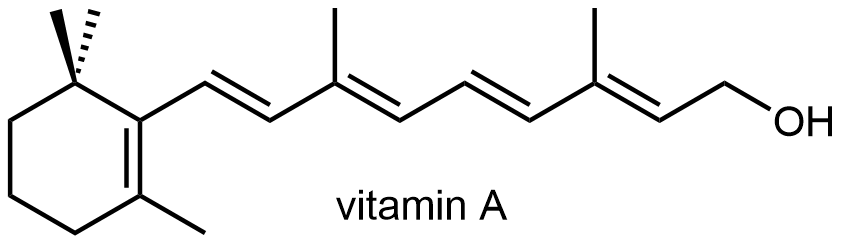
Retinol (Vitamin A) is necessary for normal juvenile growth. Vitamin A deficiency also leads to night blindness.
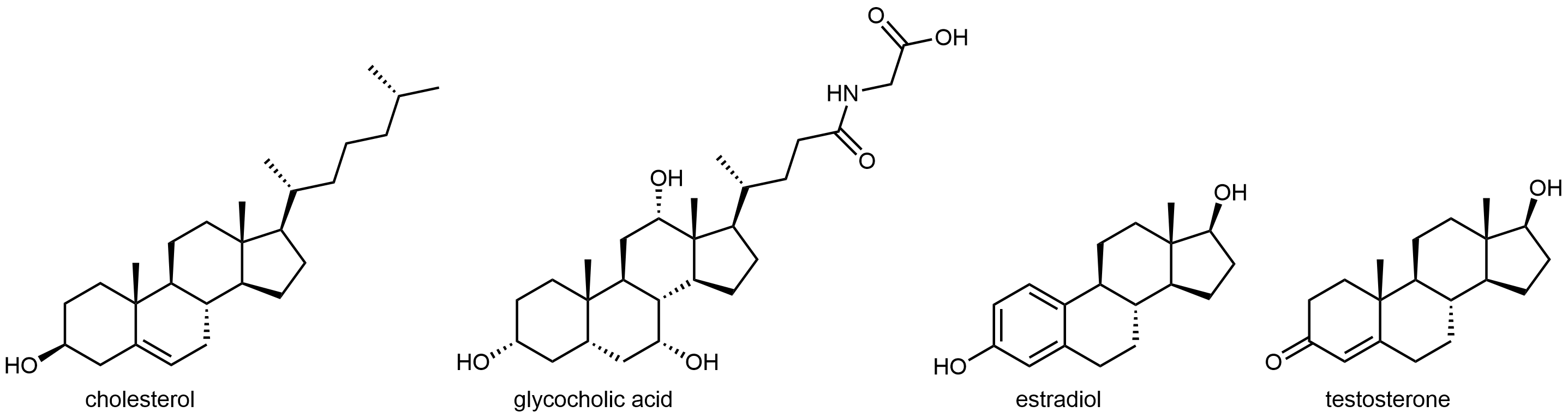
The four-ringed lipid cholesterol forms the basis for another class of lipids. Cholesterol is a building block for many important molecules used for various purposes in higher organisms. It is partially oxidized in the liver to bile salts, which act like detergents to solubilize fatty acids in foods and allow them to be absorbed by the body. Many hormones, including estrogens and testosterone, are also derived from cholesterol. The diagram below shows the structures of cholesterol, glycocholic acid (the most common bile acid), estradiol and testosterone (two well-known hormones that are derivatives of cholesterol). As you can see, all four are molecules are amphiphiles, since they have hydrophobic and hydrophilic regions.
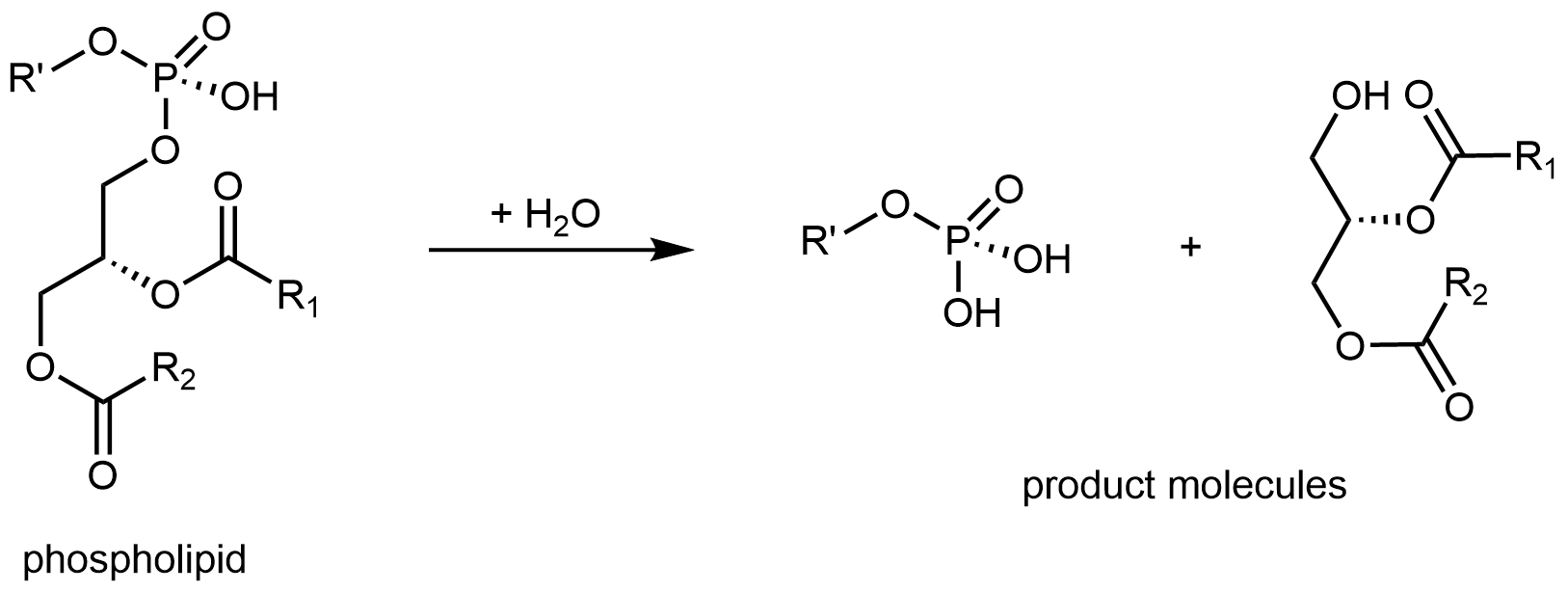
Lipids also serve a role in intracellular signaling. Some stimuli, such as the smell of bell peppers in olfactory receptor cells, activate an enzyme that breaks up phospholipids into smaller molecules. These molecules, derivatives of glycerol and fatty acids, then act on other enzymes in the cell.
Day 16 Pre-class Podia Problem: Polyfluoroalkyl Substances
The presence of per- and polyfluoroalkyl substances (PFASs) in the environment is a significant public health issue. Some sites in Wisconsin and many in Michigan and other states have been identified as being contaminated by these compounds. They’ve been dubbed “forever chemicals” because they don’t degrade naturally. See the Science article at this link for more information. The Science article says that one of the problems of PFASs is the fact that some of the molecules involved have structures that enable water and airborne droplets to carry them long distances away from their sources.
1. One of the PFASs is perfluorooctanoic acid, which is used in the manufacture of teflon polymers. (Perfluoro means that all H atoms in a hydrocarbon chain have been replaced by F atoms.) Write a Lewis structure for perfluorooctanoic acid.
2. Consider the structure of perfluorooctanoic acid and the statement above that “some of the molecules involved have structures that enable water and airborne droplets to carry them long distances.” Use models of molecular structure and intermolecular forces to construct an explanation of this fact.
3. Other perfluoroalkyl substances have longer chains of carbon atoms than perfluorooctanoic acid. In what type of body tissue would you expect to find these substances? Use models of molecular structure and intermolecular forces to answer the question and construct an explanation.
Two days before the next whole-class session, this Podia question will become live on Podia, where you can submit your answer.

