Lab 10: DATA ANALYSIS
Your data will be provided by your TAs after you complete the in-lab activities.
Complete the following steps to calculate Vmax, KM, kcat and kcat/KM for wt and mutant HCAII.
- Make a table of of initial velocities (in µM product/time) and substrate concentrations. To calculate µM product/time, you need to know: The molar absorption coefficient of PNP is 1.73 x 104 M-1cm-1 at 405 nm, and the effective path length (b) of the spectrophotometer cuvettes is 1.0 cm.
Michaelis-Menten Fit:
NOTE: The fit described here is very similar to the one from Lab 9. If you need more guidance within Prism than what is written here, refer back to the data analysis from Lab 9.
- Open GraphPad Prism and select “Enter 3 replicate values in side-by-side subcolumns,” hit “Create”
- Paste your data in. You can put replicates in adjacent columns.
- Select “Analyze”
- Under “XY analyses” select “Nonlinear regression (curve fit)” Press OK
- Under “Enzyme kinetics – Velocity as a Function of Substrate” choose “Michaelis-Menten.”
- Go into the “Compare” tab to have Prism compare the KM for wild type versus mutant.
- Go to the “Confidence” tab and select “Symmetrical (asymptotic) approximate CI” then select “Show SE of parameters.” Press OK.
- You should now have values for Vmax and KM with standard errors, a determination of statistical significance for the KM including a p-value, and a plot showing the fit.
- Also make note of the “Degrees of freedom” provided for both wt and mutant. You will need this number in a future analysis.
- Note that you also now have a plot of Vo vs [S]. You can play with the plot in Prism to make it look nice, just as you would in Excel: label axes, remove title, add error bars, etc.
kcat Calculation:
- Calculate kcat for your wt and mutant enzymes based on your Michaelis-Menten fit using Equation 3. Pay attention to units: the units of kcat are sec-1.
(3) ![]()
- You are manipulating values that all have their own associated error. You will need to report standard error values for kcat using the principles of propagation of error. Refer to the Appendix for additional help (link opens in a new tab): Appendix 2: Statistical methods used in 551 data analyses
- Once you have determined the standard error for kcat for both wild type and mutant, you can utilize a different type of Prism analysis to determine p-values.
- Open Prism and select from the far left menu “Column” (by contrast, the default is “XY”)
- Select “Enter and plot error values already calculated elsewhere” and from the dropdown menu choose “Mean, SEM, N”
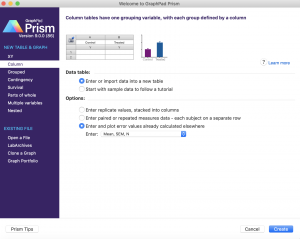
- Enter the following for wt and mutant:
- Mean: the kcat values you determined
- SEM: the standard error of the kcat values (determined by propagation of error)
- N: the degrees of freedom (determined by Prism above) + 1
- Go to “Analyze” and select, under Column analyses, “t tests (and nonparametric tests.” Press OK. (Leave all the parameters and options as the default.)
- The Results sheet should provide a p-value.
kcat /KM Calculation:
- Now take the kcat values you calculated and divide them by KM to determine the catalytic efficiency.
- You will again need to use principles of propagation of error to determine standard errors for each kcat /KM value.
- Repeat the Prism analysis above to determine a p-value for kcat /KM.

